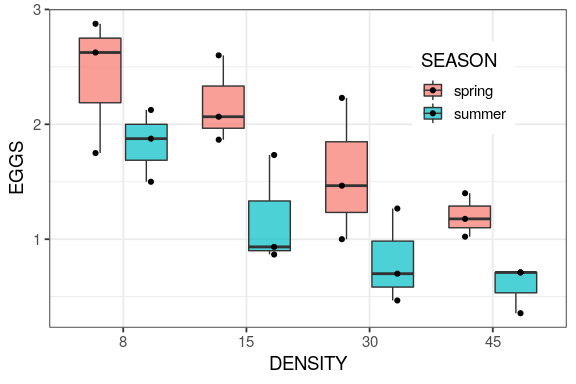
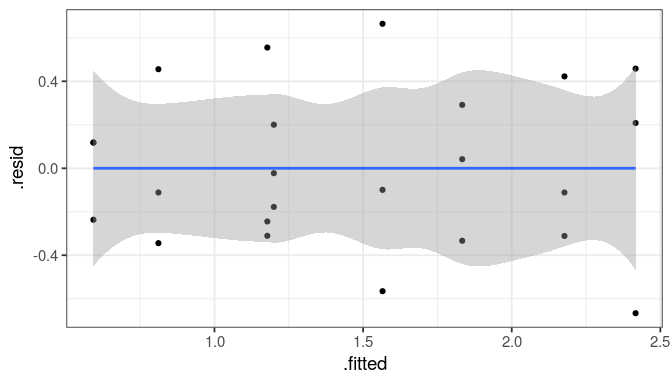
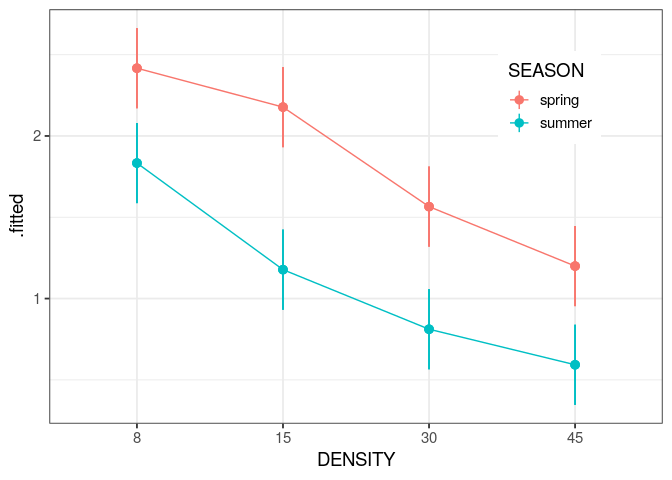
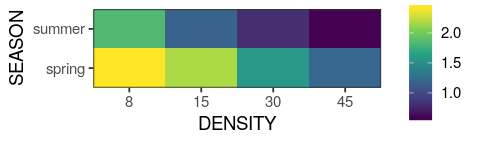
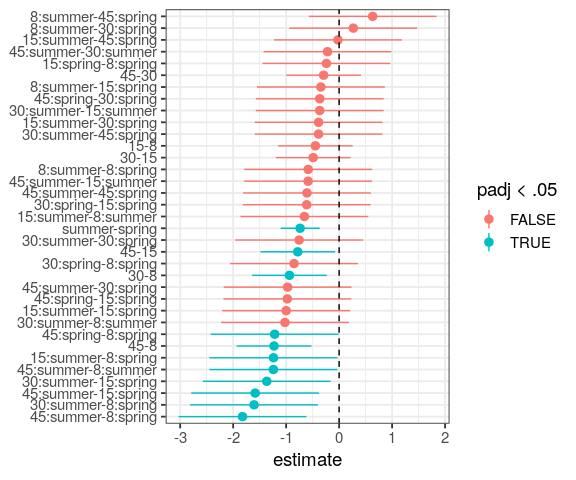
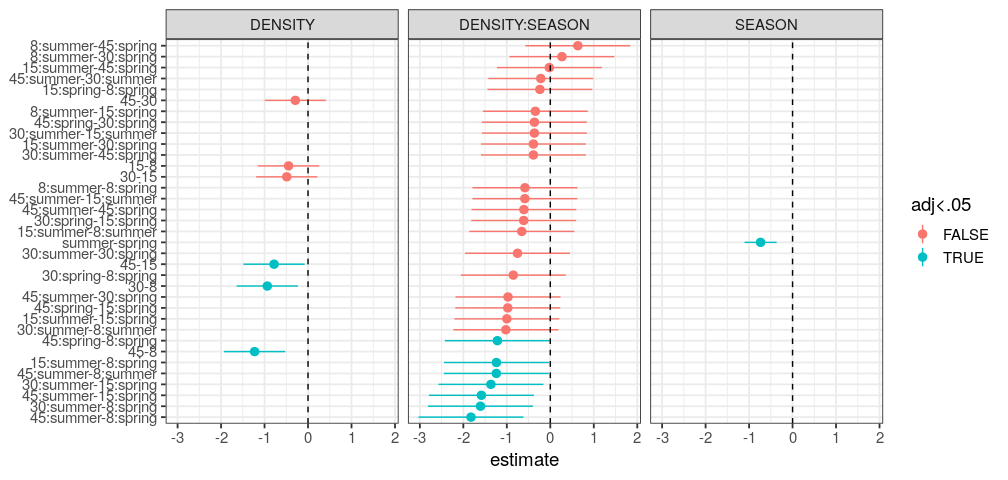
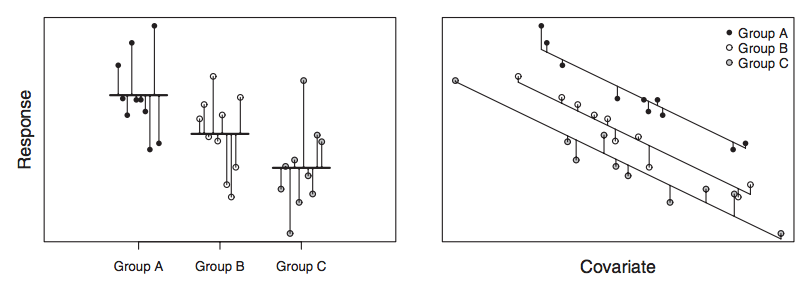
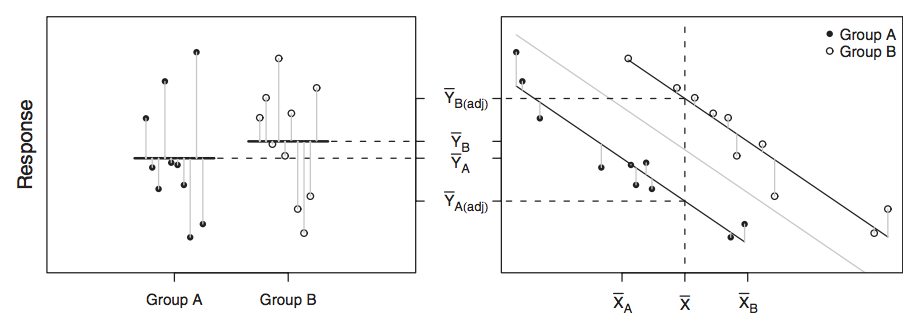
You will learn to:

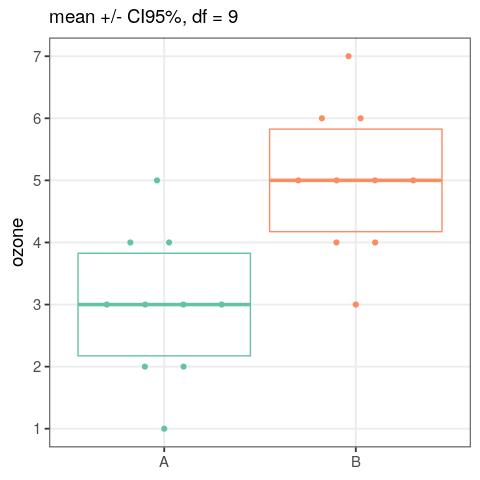
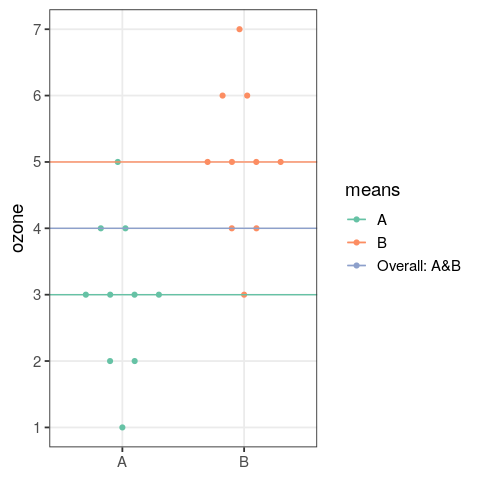
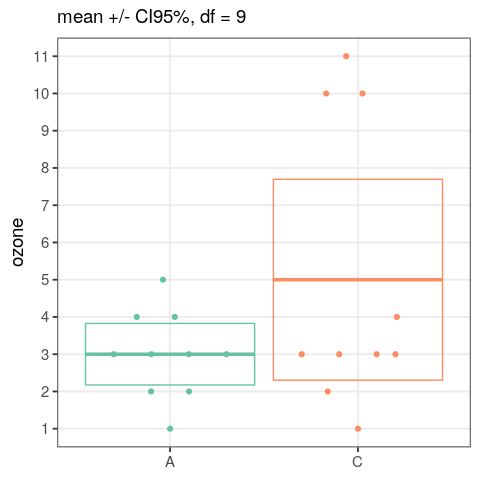
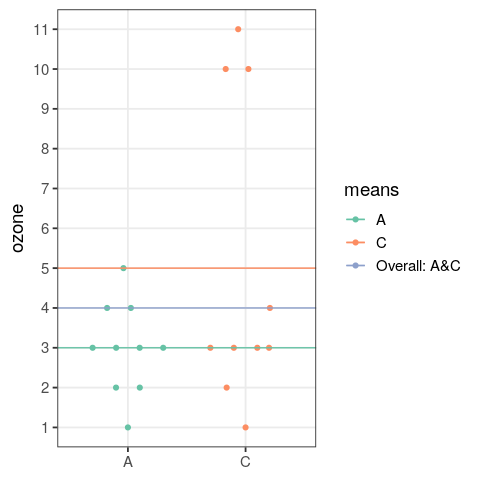
- perform an analysis of variance
- understand that it is only a linear model
- assess the output
- perform post-hoc tests
Reading

- Statistics. An introduction using R by Michael J. Crawley
- Biostatistic Design and Analysis Using R by Murray Logan (free pdf)