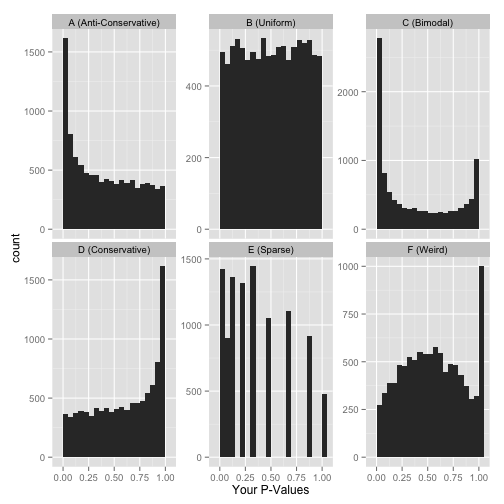
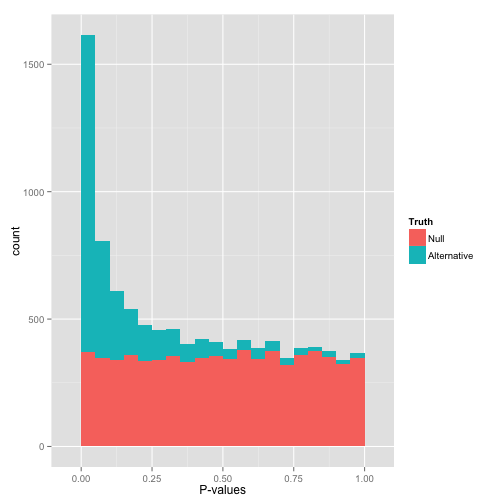
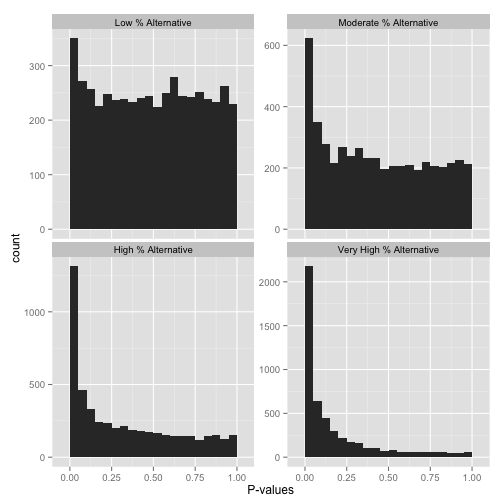
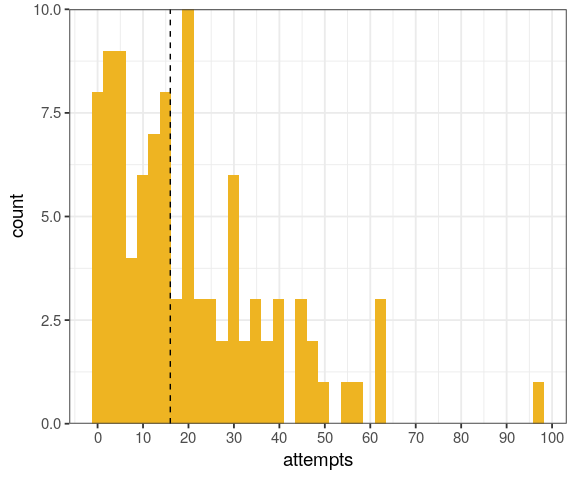
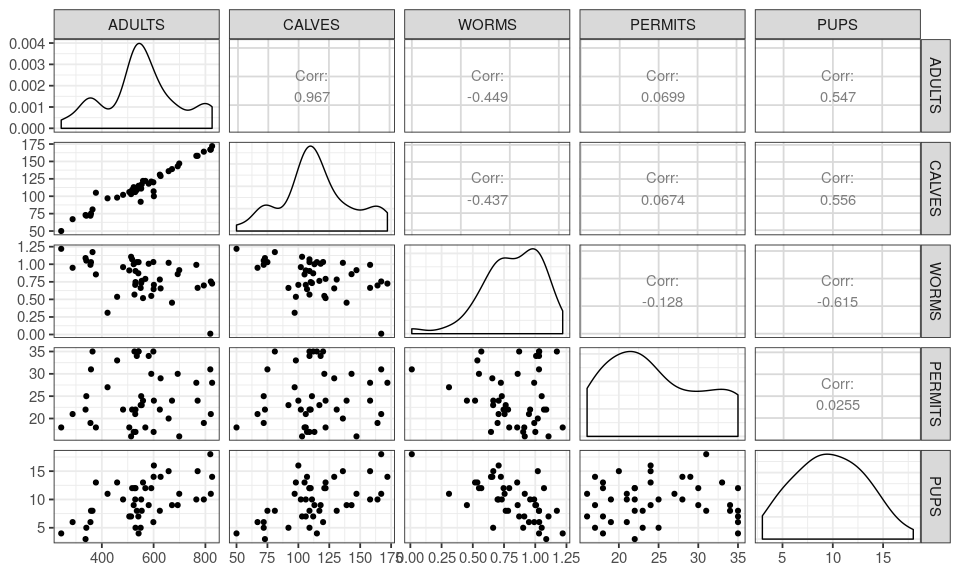
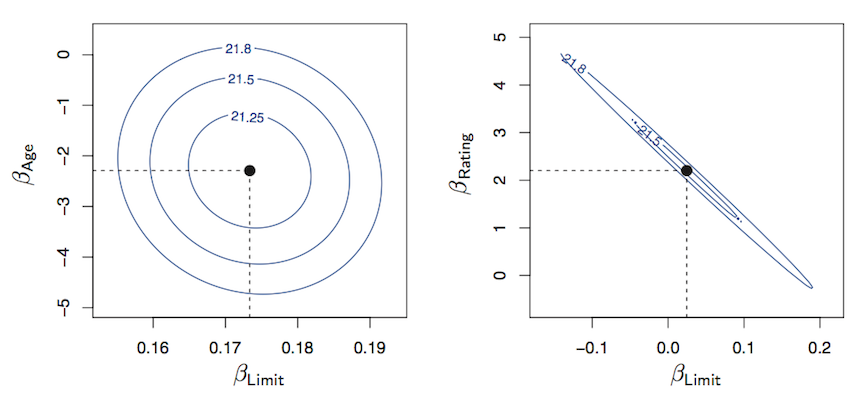
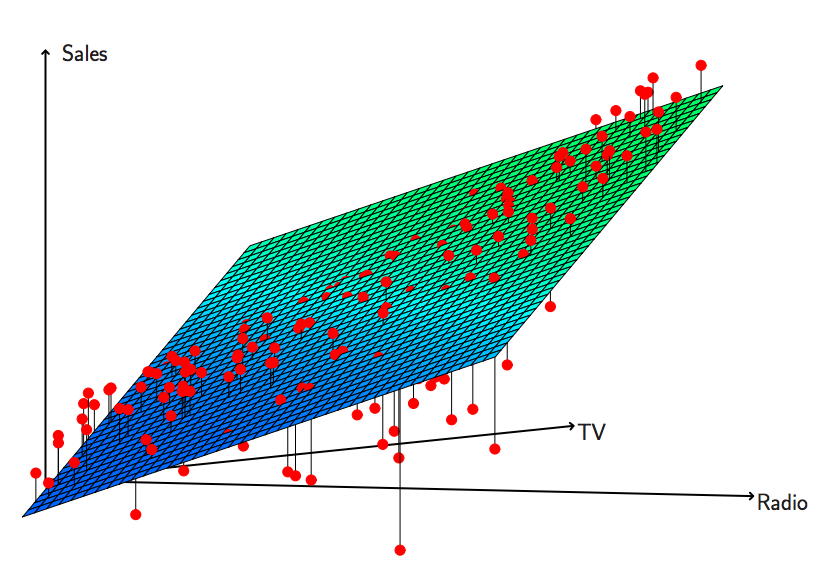
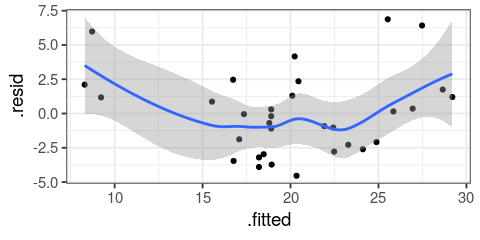
You will learn to:

- on p-values
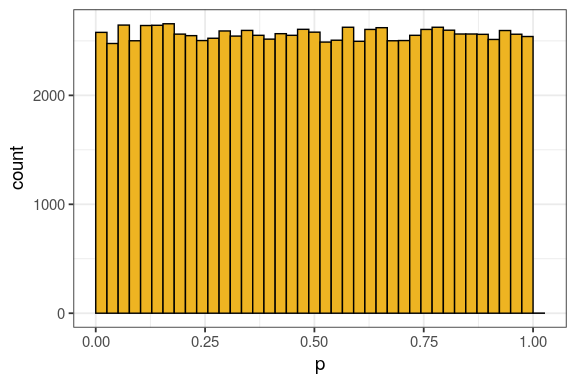
- know what a p-value actually is
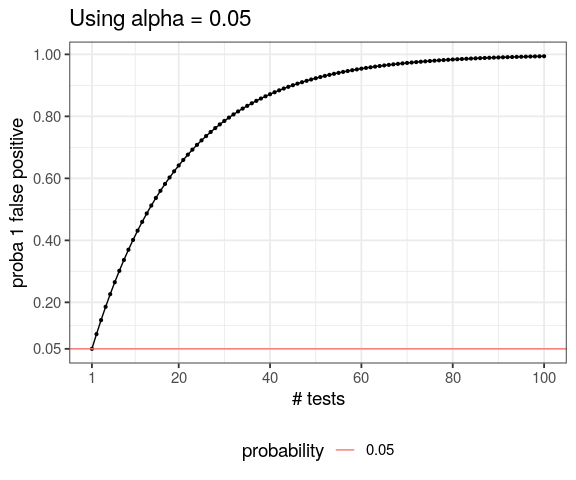
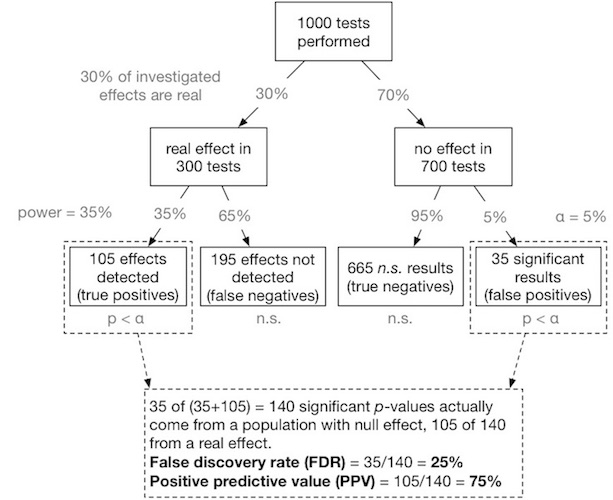
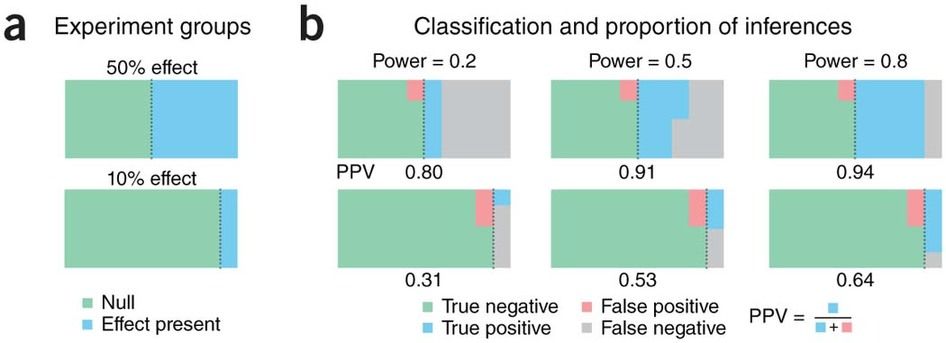
- understand necessity to correct for multiple testing
- on multiple regression
- how different from simple regression
- model simplification
- testing for predictor interaction
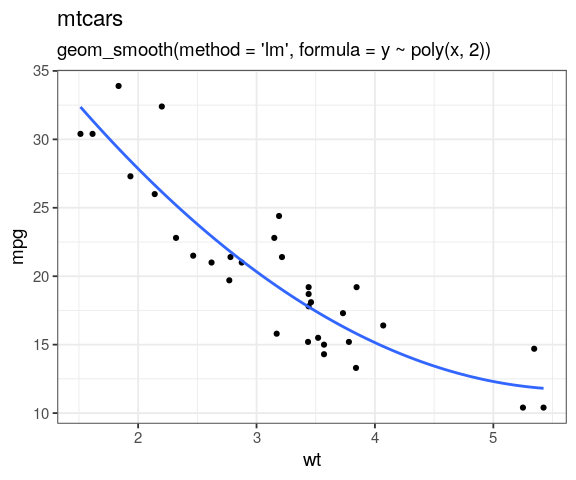
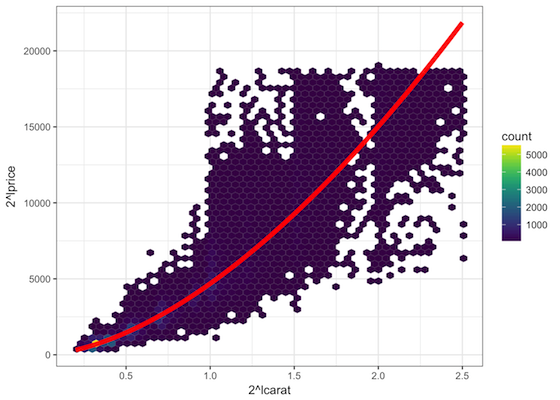
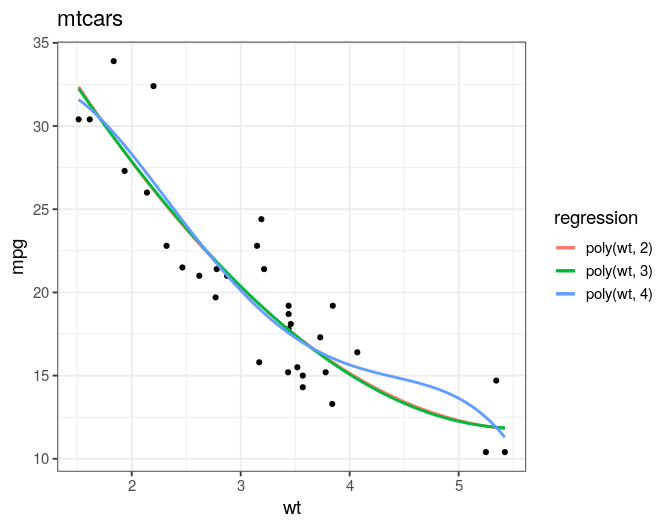
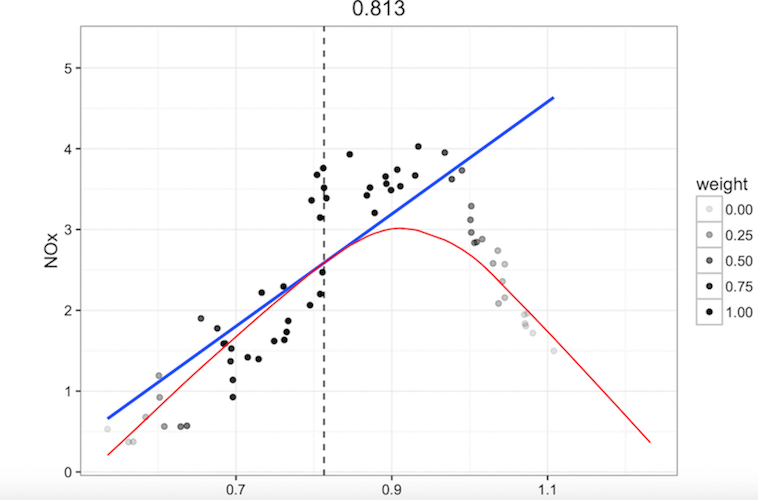
- transforming predictor to seek linearity
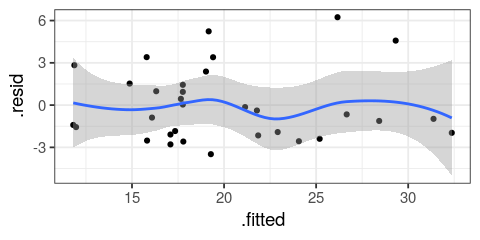
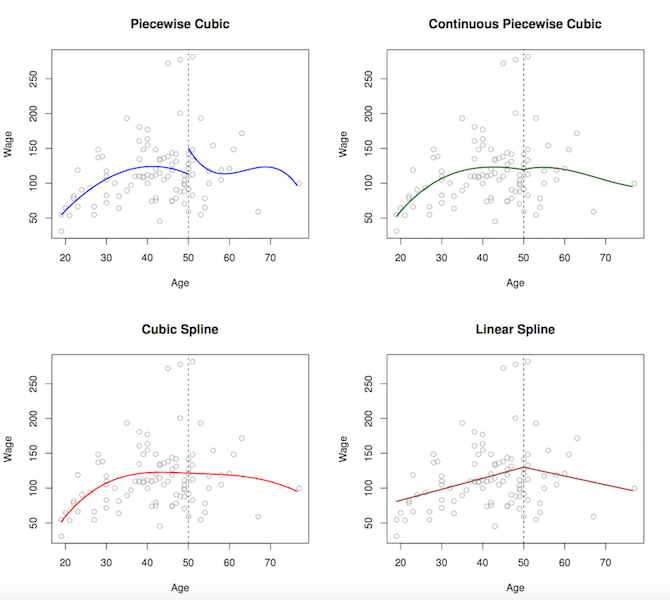
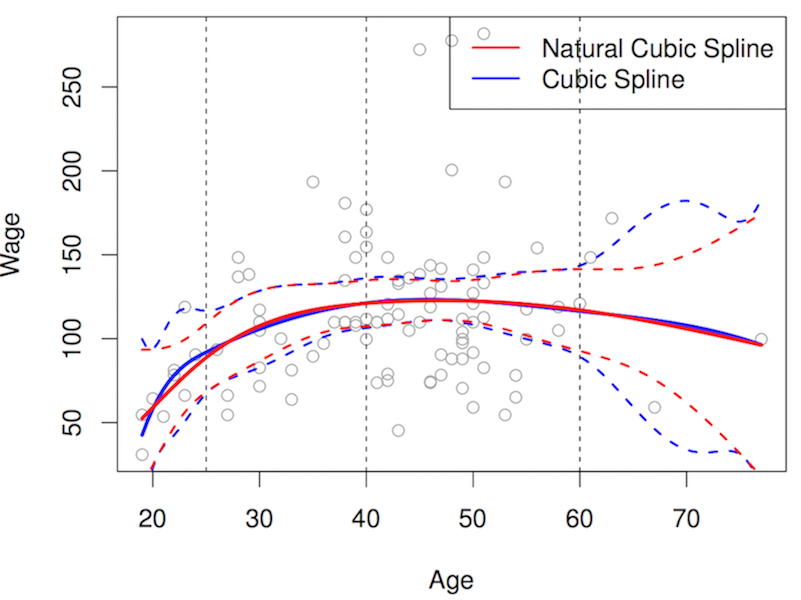
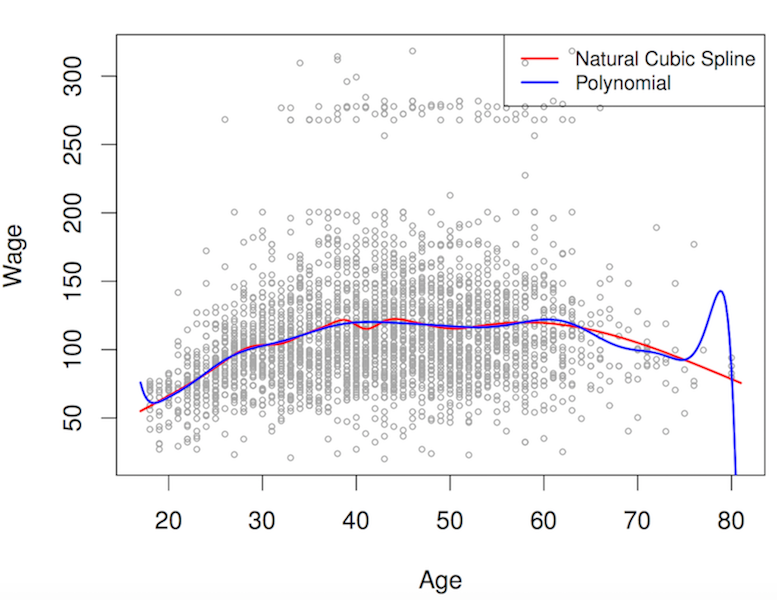
- smoothing predictors
Reading

- An Introduction to Statistical Learning by James, Witten, Hastie & Tibshirani