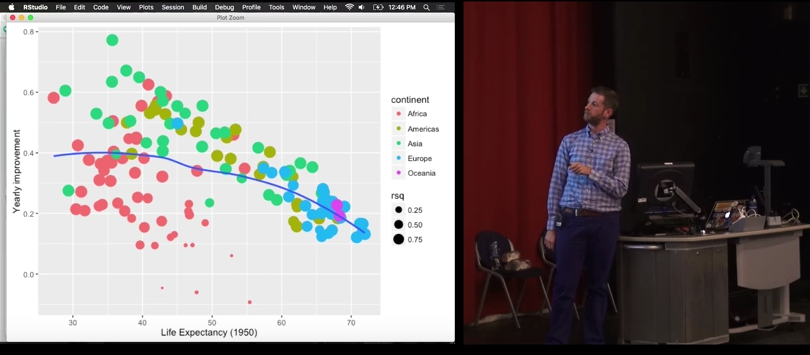
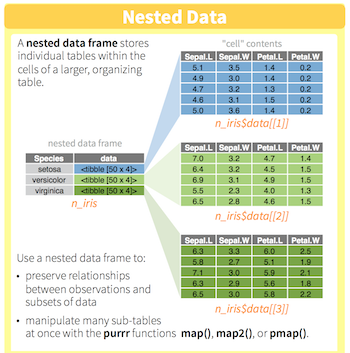
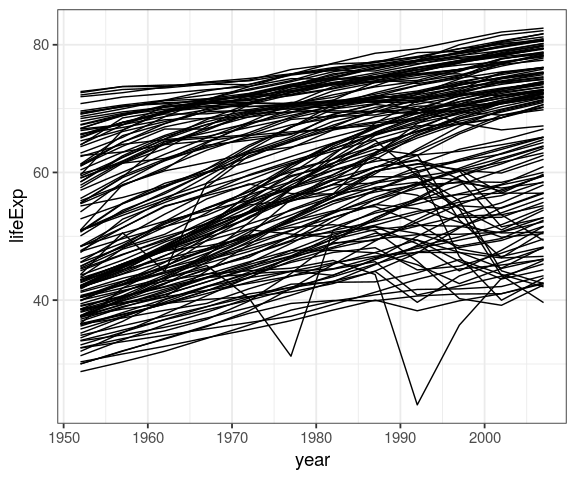
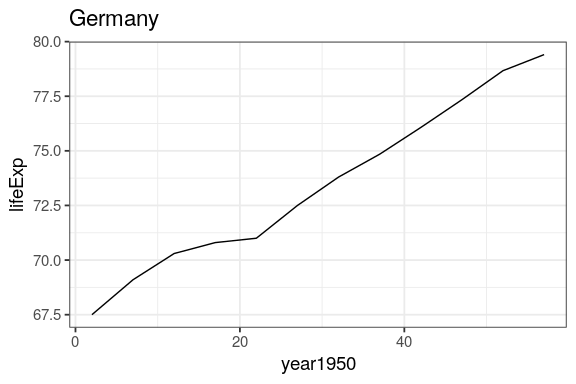
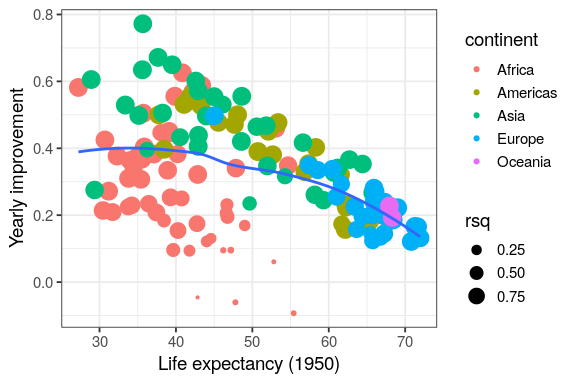
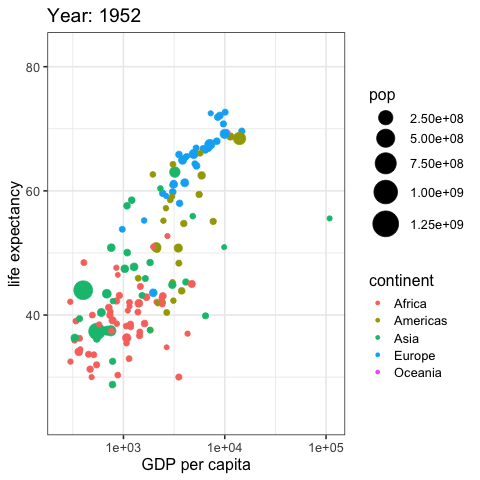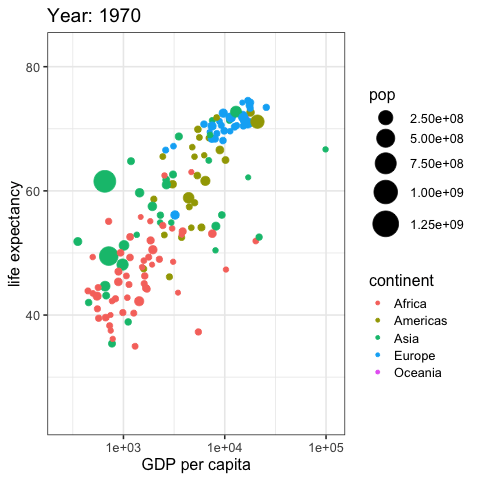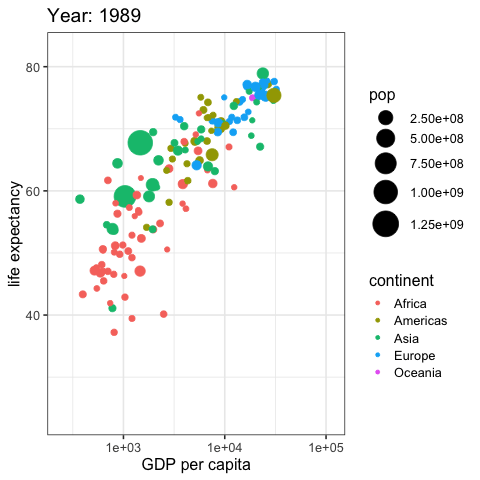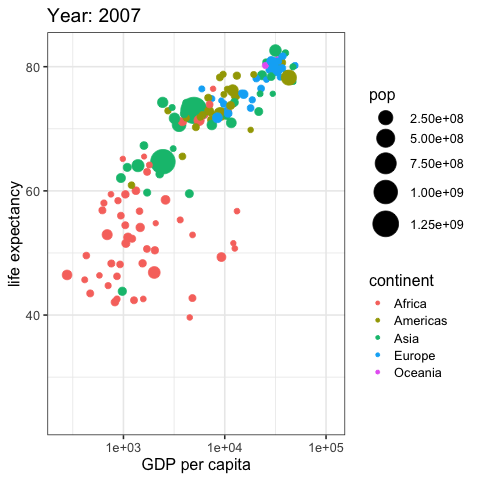
by_country %>%
filter(country == "Germany")
# A tibble: 1 x 3
continent country data
<fct> <fct> <list>
1 Europe Germany <tibble [12 × 5]>
by_country %>%
filter(country == "Germany") %>%
unnest(data) %>%
select(-country, -continent, -pop)
# A tibble: 12 x 4
year lifeExp gdpPercap year1950
<int> <dbl> <dbl> <dbl>
1 1952 67.5 7144. 2
2 1957 69.1 10188. 7
3 1962 70.3 12902. 12
4 1967 70.8 14746. 17
5 1972 71 18016. 22
6 1977 72.5 20513. 27
7 1982 73.8 22032. 32
8 1987 74.8 24639. 37
9 1992 76.1 26505. 42
10 1997 77.3 27789. 47
11 2002 78.7 30036. 52
12 2007 79.4 32170. 57