You will learn to:

- use a functional programming approach to simplify your code
- pass functions as arguments to higher order functions
- use
purrr::mapto replaceforloops - use
purrr::maptogether with nested tibbles
November 2019

purrr::map to replace for loopspurrr::map together with nested tibblesEach atomic vector contains only a single type of data
# Logical c(TRUE, FALSE, TRUE)
[1] TRUE FALSE TRUE
# double c(1, 5, 7)
[1] 1 5 7
# character
c("character", "sequence")[1] "character" "sequence"
as.*() functions)v_example <- c(1, 3, 7) str(as.character(v_example))
chr [1:3] "1" "3" "7"
v_example <- c(1, 3) str(v_example)
num [1:2] 1 3
str(c(v_example, "seven"))
chr [1:3] "1" "3" "seven"
Adapted from the tutorial of Jennifer Bryan
my_list <- list(1, 3, "seven") str(my_list)
List of 3 $ : num 1 $ : num 3 $ : chr "seven"
is.vector(my_list)
[1] TRUE
is.atomic(my_list)
[1] FALSE
Adapted from the tutorial of Jennifer Bryan
my_function <- function(my_argument) {
my_argument + 1
}is defined in the global environment
ls()
[1] "my_function"
ls.str()
my_function : function (my_argument)
is reusable
my_function(2)
[1] 3
(function(x) { x + 1 })(2)
[1] 3
Does not alter the global environment
ls()
[1] "my_function"
# remove the previous my_function to convince you
rm(my_function)
(function(x) { x + 1 })(2)
[1] 3
ls()
character(0)


purrr enhances R’s functional programming toolkit by providing a complete and consistent set of tools for working with functions and vectors (purrr overview on github page)
functional programming is a programming paradigm – a style of building the structure and elements of computer programs – that treats computation as the evaluation of mathematical functions and avoids changing-state and mutable data Wikipedia
FOR EACH x DO f
put_on functionput_on(x, antenna)

legos

antenna

lego with antenna

legos

antenna

legos with antennas
out <- vector("list", length(legos))
for (i in seq_along(legos)) {
out[[i]] <- put_on(legos[[i]], antenna)
}
outantennate <- function(x) put_on(x, antenna) map(legos, antennate)
a data frame is a list
is.list(mtcars)
[1] TRUE
Each column represents an element of the list i.e. a data frame is a list of columns

Calculate the mean of each column of the mtcars dataset.
03:00
purrr::map()
map(mtcars, mean) %>% str()
List of 11 $ mpg : num 20.1 $ cyl : num 6.19 $ disp: num 231 $ hp : num 147 $ drat: num 3.6 $ wt : num 3.22 $ qsec: num 17.8 $ vs : num 0.438 $ am : num 0.406 $ gear: num 3.69 $ carb: num 2.81
for loops
means <- vector("list", ncol(mtcars))
for (i in seq_along(mtcars)) {
means[i] <- mean(mtcars[[i]])
}
# need to manually add names
names(means) <- names(mtcars)
means %>%
str()List of 11 $ mpg : num 20.1 $ cyl : num 6.19 $ disp: num 231 $ hp : num 147 $ drat: num 3.6 $ wt : num 3.22 $ qsec: num 17.8 $ vs : num 0.438 $ am : num 0.406 $ gear: num 3.69 $ carb: num 2.81
for_loop <- function(x) {
res <- c()
for (i in seq_len(x)) {
res[i] <- i
}
}for_loop <- function(x) {
res <- vector(mode = "integer",
length = x)
for (i in seq_len(x)) {
res[i] <- i
}
}library(Rcpp)
cppFunction("NumericVector rcpp(int x) {
NumericVector res(x);
for (int i=0; i < x; i++) {
res[i] = i;
}
}")

legos

antenna

map(legos, antennate)
purrr::map() is type stablemap(YOUR_LIST, YOUR_FUNCTION)base::apply() vs purrr::map()apply() family of function is inconsistent and some members are not type stable.lapply()apply() family still useful to avoid dependencies (package development)| function | call | input | output |
|---|---|---|---|
apply() |
apply(X, MARGIN, FUN, ...) |
array | vector or array or list |
lapply() |
lapply(X, FUN, ...) |
list | list |
sapply() |
sapply(X, FUN, ...) |
list | vector or array or list |
vapply() |
vapply(X, FUN, FUN.VALUE, ...) |
list | specified |
base::apply() vs purrr::map() apply()
apply(mtcars, 2, mean) %>% str()
Named num [1:11] 20.09 6.19 230.72 146.69 3.6 ... - attr(*, "names")= chr [1:11] "mpg" "cyl" "disp" "hp" ...
lapply()
lapply(mtcars, mean) %>% str()
List of 11 $ mpg : num 20.1 $ cyl : num 6.19 $ disp: num 231 $ hp : num 147 $ drat: num 3.6 $ wt : num 3.22 $ qsec: num 17.8 $ vs : num 0.438 $ am : num 0.406 $ gear: num 3.69 $ carb: num 2.81
base::apply() vs purrr::map()Let’s create the following tibbles:
tib_1 <- tibble(a = 1:3, b = 4:6, c = 7:9) tib_2 <- tibble(a = 1:3, b = 4:6, c = 11:13)
troublemaker functiontroublemaker <- function(x) {
if (any(x > 10)) return(NULL)
sum(x)
}
troublemaker_chr <- function(x) {
if (any(x > 10)) return(as.character(sum(x)))
sum(x)
}Define some troublemaker functions:
troublemaker() returns NULLtroublemaker_chr() coerces the sum to a characterbase::apply() vs purrr::map()troublemaker example
troublemaker(1:3)
[1] 6
troublemaker(11:13)
NULL
troublemaker_chr(1:3)
[1] 6
troublemaker_chr(11:13)
[1] "36"
lapply()
lapply(tib_1, troublemaker) %>% str()
List of 3 $ a: int 6 $ b: int 15 $ c: int 24
lapply(tib_2, troublemaker) %>% str()
List of 3 $ a: int 6 $ b: int 15 $ c: NULL
lapply(tib_2, troublemaker_chr) %>% str()
List of 3 $ a: int 6 $ b: int 15 $ c: chr "36"
map()
map(tib_1, troublemaker) %>% str()
List of 3 $ a: int 6 $ b: int 15 $ c: int 24
map(tib_2, troublemaker) %>% str()
List of 3 $ a: int 6 $ b: int 15 $ c: NULL
map(tib_2, troublemaker_chr) %>% str()
List of 3 $ a: int 6 $ b: int 15 $ c: chr "36"
base::apply() vs purrr::map()apply()
apply(tib_1, 2, troublemaker) %>% str()
Named int [1:3] 6 15 24 - attr(*, "names")= chr [1:3] "a" "b" "c"
apply(tib_2, 2, troublemaker) %>% str()
List of 3 $ a: int 6 $ b: int 15 $ c: NULL
apply(tib_2, 2, troublemaker_chr) %>% str()
Named chr [1:3] "6" "15" "36" - attr(*, "names")= chr [1:3] "a" "b" "c"

In addition to the confusing margin argument, apply() simplifies the output value: read the help ?apply.
If each call to FUN returns a vector of length n, then apply returns an array of dimension c(n, dim(X)[MARGIN]) if n > 1. If n equals 1, apply returns a vector if MARGIN has length 1 and an array of dimension dim(X)[MARGIN] otherwise. If n is 0, the result has length 0 but not necessarily the ‘correct’ dimension.
If the calls to FUN return vectors of different lengths, apply returns a list of length prod(dim(X)[MARGIN]) with dim set to MARGIN if this has length greater than one.
In all cases the result is coerced by as.vector to one of the basic vector types before the dimensions are set, so that (for example) factor results will be coerced to a character array. value definition in?apply
purrr::map()map() variants
purrr provides variants coercing the output to the desired output. Generates an error if the mapped function produces an unexpected output.
map_dbl()
map_dbl(tib_1, troublemaker) %>% str()
Named num [1:3] 6 15 24 - attr(*, "names")= chr [1:3] "a" "b" "c"
map_dbl(tib_2, troublemaker) %>% str()
Result 3 must be a single double, not NULL of length 0
map_dbl(tib_2, troublemaker_chr) %>% str()
Error: Can't coerce element 3 from a character to a double
base::apply() vs purrr::map()purrr::map() family of functionsmap() is the general function and close to base::lapply()map() introduces shortcuts (absent in lapply())map_lgl()map_int()map_dbl()map_chr()map_df()purrr::map() exampleLet’s split the mtcars dataset by each value of cylinder
spl_mtcars <- group_split(mtcars, mtcars$cyl) str(spl_mtcars, max.level = 1)
List of 3 $ :Classes 'tbl_df', 'tbl' and 'data.frame': 11 obs. of 12 variables: $ :Classes 'tbl_df', 'tbl' and 'data.frame': 7 obs. of 12 variables: $ :Classes 'tbl_df', 'tbl' and 'data.frame': 14 obs. of 12 variables: - attr(*, "ptype")=Classes 'tbl_df', 'tbl' and 'data.frame': 0 obs. of 12 variables: ..- attr(*, "groups")=Classes 'tbl_df', 'tbl' and 'data.frame': 3 obs. of 2 variables: .. ..- attr(*, ".drop")= logi TRUE
From R for Data Science
mtcars dataset we can fit a linear model to explain the miles per gallon (mpg) by the weight (wt) using:lm(mpg ~ wt, data = mtcars)
Call:
lm(formula = mpg ~ wt, data = mtcars)
Coefficients:
(Intercept) wt
37.285 -5.344 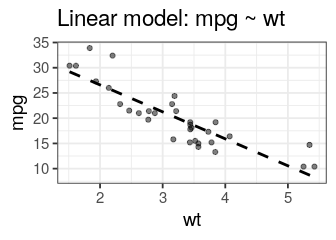
lm outputs complex objectsbase::summary() summaries the model:
lm(mpg ~ wt, data = mtcars) %>% summary()
Call:
lm(formula = mpg ~ wt, data = mtcars)
Residuals:
Min 1Q Median 3Q Max
-4.5432 -2.3647 -0.1252 1.4096 6.8727
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 37.2851 1.8776 19.858 < 2e-16 ***
wt -5.3445 0.5591 -9.559 1.29e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.046 on 30 degrees of freedom
Multiple R-squared: 0.7528, Adjusted R-squared: 0.7446
F-statistic: 91.38 on 1 and 30 DF, p-value: 1.294e-10purrr::map() example05:00
So for the 3 cyl groups:
mpg ~ wtmap(YOUR_LIST, YOUR_FUNCTION)YOUR_LIST = spl_mtcarsYOUR_FUNCTION can be an anonymous function (declared on the fly)map(spl_mtcars, function(x) lm(mpg ~ wt, data = x))
[[1]]
Call:
lm(formula = mpg ~ wt, data = x)
Coefficients:
(Intercept) wt
39.571 -5.647
[[2]]
Call:
lm(formula = mpg ~ wt, data = x)
Coefficients:
(Intercept) wt
28.41 -2.78
[[3]]
Call:
lm(formula = mpg ~ wt, data = x)
Coefficients:
(Intercept) wt
23.868 -2.192 purrr::map() examplemap(spl_mtcars,
function(x) lm(mpg ~ wt, data = x))
[[1]]
Call:
lm(formula = mpg ~ wt, data = x)
Coefficients:
(Intercept) wt
39.571 -5.647
[[2]]
Call:
lm(formula = mpg ~ wt, data = x)
Coefficients:
(Intercept) wt
28.41 -2.78
[[3]]
Call:
lm(formula = mpg ~ wt, data = x)
Coefficients:
(Intercept) wt
23.868 -2.192 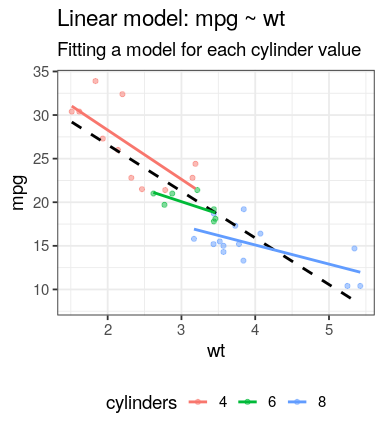
base::summary() generates a list
lm(mpg ~ wt, data = mtcars) %>% summary() %>% str(max.level = 1, give.attr = FALSE)
List of 11 $ call : language lm(formula = mpg ~ wt, data = mtcars) $ terms :Classes 'terms', 'formula' language mpg ~ wt $ residuals : Named num [1:32] -2.28 -0.92 -2.09 1.3 -0.2 ... $ coefficients : num [1:2, 1:4] 37.285 -5.344 1.878 0.559 19.858 ... $ aliased : Named logi [1:2] FALSE FALSE $ sigma : num 3.05 $ df : int [1:3] 2 30 2 $ r.squared : num 0.753 $ adj.r.squared: num 0.745 $ fstatistic : Named num [1:3] 91.4 1 30 $ cov.unscaled : num [1:2, 1:2] 0.38 -0.1084 -0.1084 0.0337
fit_summary <- summary(lm(mpg ~ wt, data = mtcars)) fit_summary$r.squared
[1] 0.7528328
purrr::map() example%>%)spl_mtcars %>% map(function(x) lm(mpg ~ wt, data = x)) %>% map(summary) %>% map(function(x) x$r.squared)
[[1]] [1] 0.5086326 [[2]] [1] 0.4645102 [[3]] [1] 0.4229655

The code above can be simplified using shortcuts provided by purrr
purrr::map() shortcuts~.x to refer to the current list element (.x represents the argument of the anonymous function)map(YOUR_LIST, function(x) lm(mpg ~ wt, data = x)) # is equivalent to: map(YOUR_LIST, ~ lm(mpg ~ wt, data = .x))
spl_mtcars %>% map(function(x) lm(mpg ~ wt, data = x)) %>% map(summary) %>% map(function(x) x$r.squared)
spl_mtcars %>% map(~lm(mpg ~ wt, data = .x)) %>% map(summary) %>% map(~.x$r.squared)
purrr::map() shortcutsUse a string to extract named components
map(spl_mtcars, "mpg")
[[1]] [1] 22.8 24.4 22.8 32.4 30.4 33.9 21.5 27.3 26.0 30.4 21.4 [[2]] [1] 21.0 21.0 21.4 18.1 19.2 17.8 19.7 [[3]] [1] 18.7 14.3 16.4 17.3 15.2 10.4 10.4 14.7 15.5 15.2 13.3 19.2 15.8 15.0
Use a number to extract by index
map(spl_mtcars, 1)
[[1]] [1] 22.8 24.4 22.8 32.4 30.4 33.9 21.5 27.3 26.0 30.4 21.4 [[2]] [1] 21.0 21.0 21.4 18.1 19.2 17.8 19.7 [[3]] [1] 18.7 14.3 16.4 17.3 15.2 10.4 10.4 14.7 15.5 15.2 13.3 19.2 15.8 15.0
purrr::map() shortcutsspl_mtcars %>% map(function(x) lm(mpg ~ wt, data = x)) %>% map(summary) %>% map(function(x) x$r.squared)
[[1]] [1] 0.5086326 [[2]] [1] 0.4645102 [[3]] [1] 0.4229655
spl_mtcars %>% map(~lm(mpg ~ wt, data = .x)) %>% map(summary) %>% map(~.x$r.squared)
[[1]] [1] 0.5086326 [[2]] [1] 0.4645102 [[3]] [1] 0.4229655
spl_mtcars %>%
map(~lm(mpg ~ wt, data = .x)) %>%
map(summary) %>%
map("r.squared")[[1]] [1] 0.5086326 [[2]] [1] 0.4645102 [[3]] [1] 0.4229655
purrr::map_*() variantsmap()
spl_mtcars %>%
map(~lm(mpg ~ wt, data = .x)) %>%
map(summary) %>%
map("r.squared") %>%
str()
List of 3 $ : num 0.509 $ : num 0.465 $ : num 0.423
map_dbl()
spl_mtcars %>%
map(~lm(mpg ~ wt, data = .x)) %>%
map(summary) %>%
map_dbl("r.squared") %>%
str()
num [1:3] 0.509 0.465 0.423
purrr::map() and mutliple arguments
You can use purrr::map() shortcuts to pass additional (constant) arguments
antennate <- function(x) put_on(x, antenna) map(legos, antennate)
map(legos, ~ put_on(.x, antenna))
map(legos, put_on, antenna)

map() for a single list

map(legos, antennate)
map2() for two lists

enhair <- function(x, y) put_on(x, y) map2(legos, hairs, enhair)
pmap(.l, .f, ...)
.l)..1, ..2, ..3, …pmap() example
my_list <- list(
list(1, 5, 7),
list(6, 10, 9),
list(1, 2, 0.5)
)
pmap(my_list,
function(a, b, c) {seq(from = a, to = b, by = c)}
) %>%
str()List of 3 $ : num [1:6] 1 2 3 4 5 6 $ : num [1:3] 5 7 9 $ : num [1:5] 7 7.5 8 8.5 9
pmap(my_list, ~seq(from = ..1, to = ..2, by = ..3)) %>% str()
List of 3 $ : num [1:6] 1 2 3 4 5 6 $ : num [1:3] 5 7 9 $ : num [1:5] 7 7.5 8 8.5 9
A function might be called for its side-effects:
walk family of functionwalk(), walk2(), pwalk()%>%)dplyr
tibble introduces list-columnsdplyrgroup_nest()purrr::map()
tibble(numbers = 1:8,
my_list = list(a = c("a", "b"), b = 2.56,
c = c("a", "b"), d = rep(TRUE, 4),
d = 2:3, e = 4:6, f = "Z", g = 1:4))
# A tibble: 8 x 2
numbers my_list
<int> <named list>
1 1 <chr [2]>
2 2 <dbl [1]>
3 3 <chr [2]>
4 4 <lgl [4]>
5 5 <int [2]>
6 6 <int [3]>
7 7 <chr [1]>
8 8 <int [4]> mtcars %>% group_nest(cyl)
# A tibble: 3 x 2
cyl data
<dbl> <list>
1 4 <tibble [11 × 10]>
2 6 <tibble [7 × 10]>
3 8 <tibble [14 × 10]>mtcars %>%
group_nest(cyl) %>%
mutate(model = map(data, ~lm(mpg ~ wt, data = .x)),
summary = map(model, summary),
r_squared = map_dbl(summary, "r.squared"))
# A tibble: 3 x 5
cyl data model summary r_squared
<dbl> <list> <list> <list> <dbl>
1 4 <tibble [11 × 10]> <lm> <smmry.lm> 0.509
2 6 <tibble [7 × 10]> <lm> <smmry.lm> 0.465
3 8 <tibble [14 × 10]> <lm> <smmry.lm> 0.423dplyr, tidyr, tibble, purrr and broom nicely work togetherMake your pure #rstats functions purr with purrr, a new package for functional programming: http://t.co/91Efuz0txk
— Hadley Wickham (@hadleywickham) 29 septembre 2015
A function is called “pure” if all its inputs are declared as inputs - none of them are hidden - and likewise all its outputs are declared as outputs Kris Jenkins
start <- 10
impure <- function(x) {
print(start)
x + start
}
result <- impure(2)
[1] 10
result
[1] 12
pure <- function(x, start) {
x + start
}
result <- pure(2, start)
result
[1] 12
log() has side-effectspurrr::safely() to catch every outputlog()
(res <- log(10))
[1] 2.302585
res <- log("a")
Error in log("a"): non-numeric argument to mathematical function
res
[1] 2.302585
log()
safe_log <- purrr::safely(log) (res <- safe_log(10))
$result [1] 2.302585 $error NULL
res <- safe_log("a")
res
$result
NULL
$error
<simpleError in .Primitive("log")(x, base): non-numeric argument to mathematical function>Have a look at the purrr cheatsheet
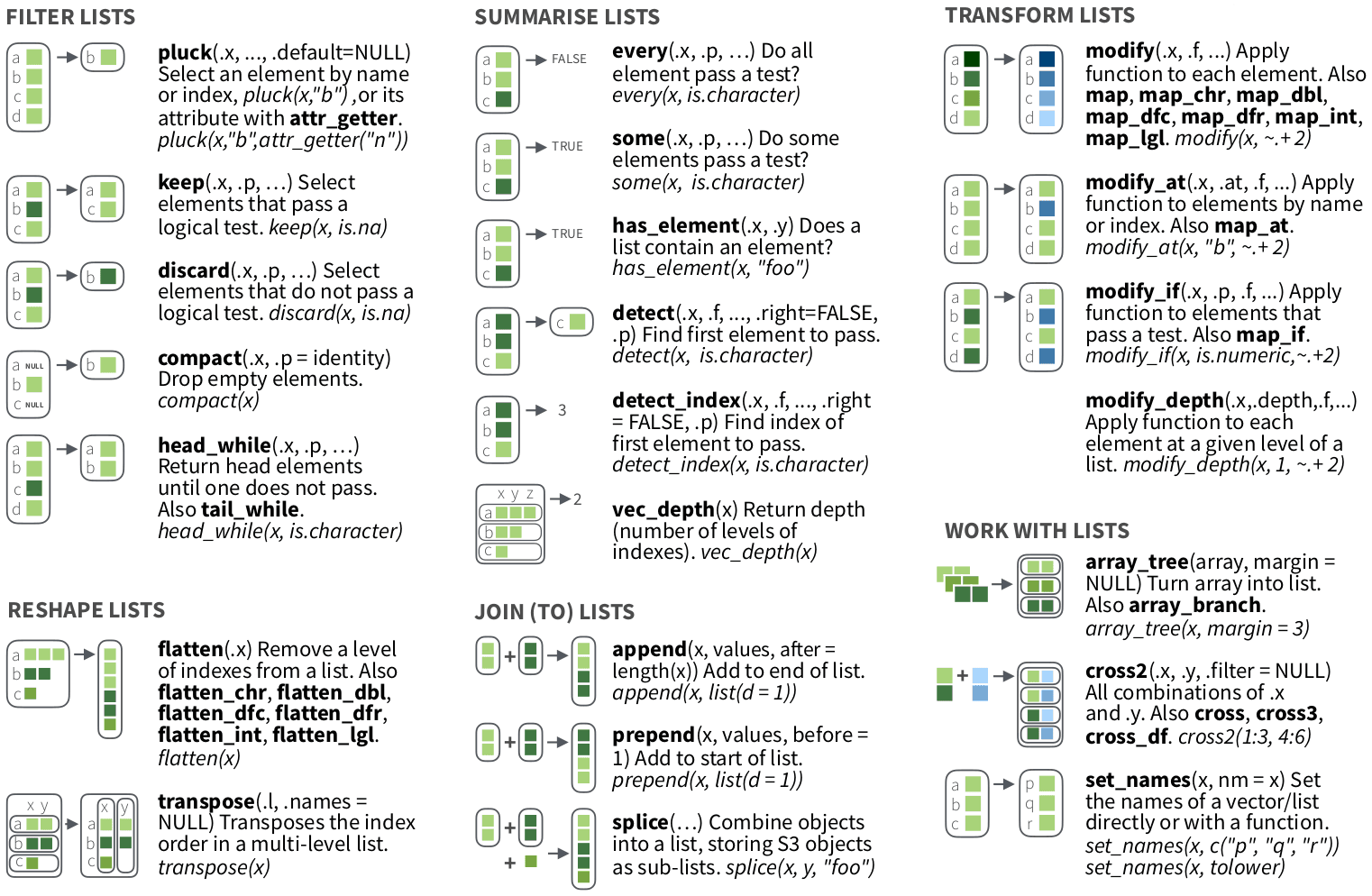
map() or walk()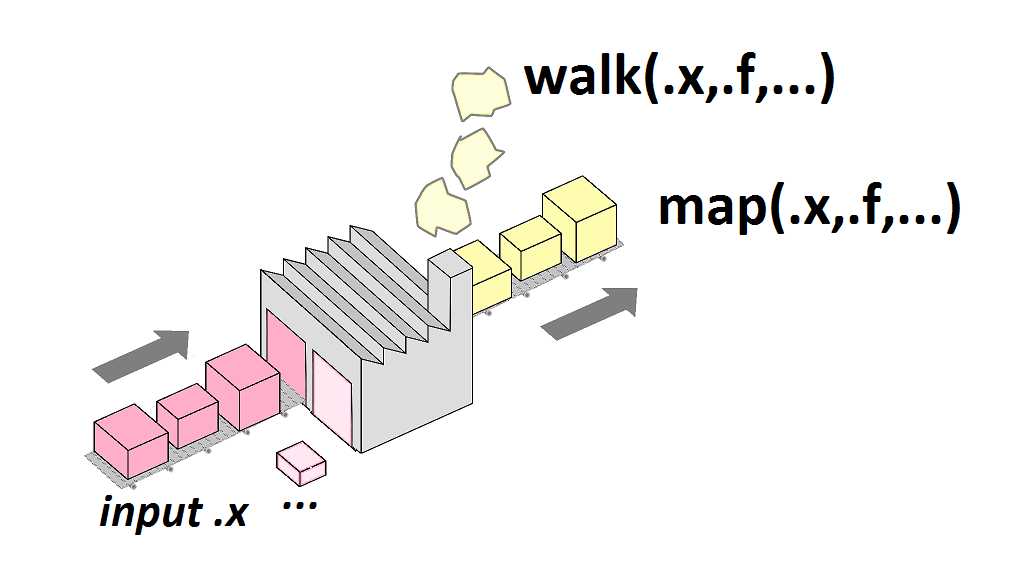
map2() or walk2()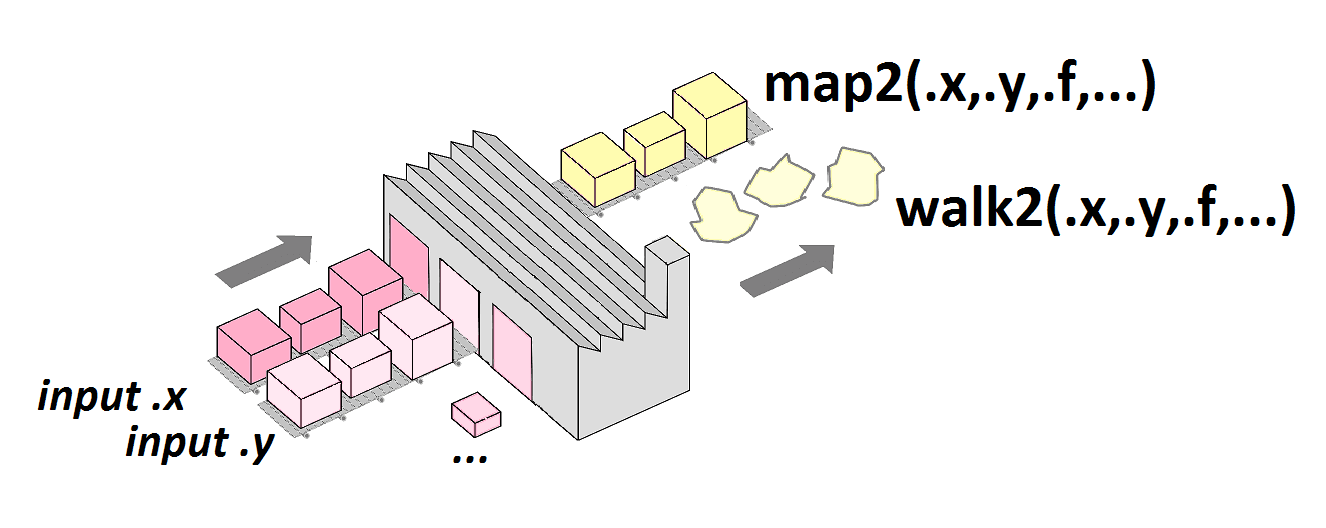
pmap()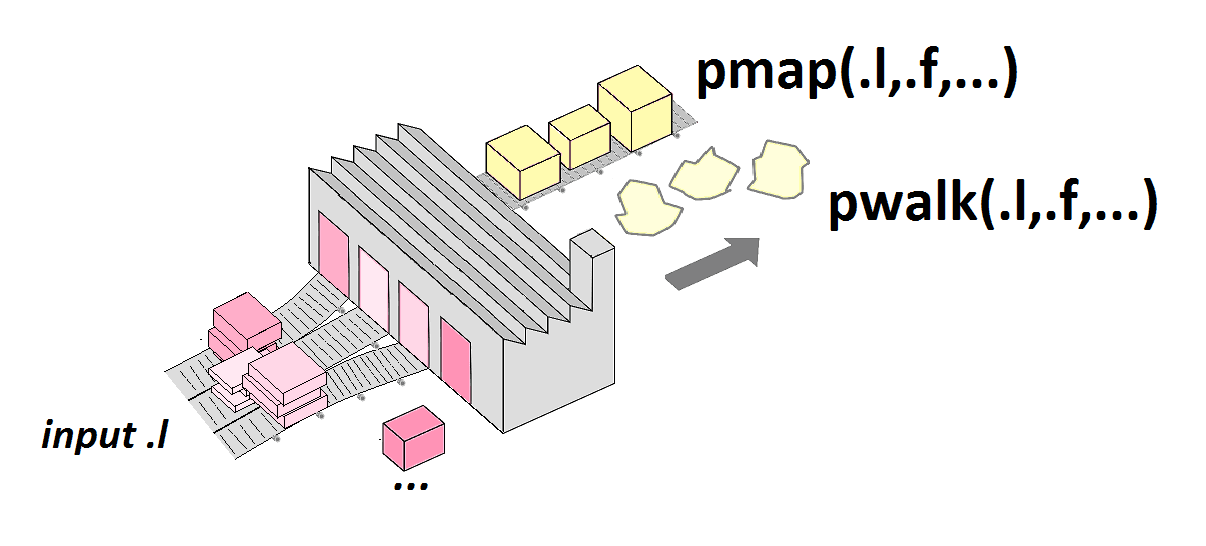
Pictures from Lise Vaudor’s blog

Don’t overmap functions!
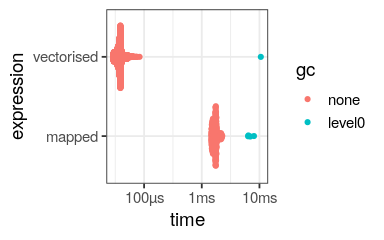
Use map only if required (non vectorised function)
nums <- sample(1:10,
size = 1000,
replace = TRUE)
log_vec <- log(nums)
log_map <- map_dbl(nums, log)
identical(log_vec, log_map)
[1] TRUE
bench::mark( vectorised = log(nums), mapped = map_dbl(nums, log)) %>% autoplot()