You will learn:

- what is a model?
model categories depending on data
- perform linear models
- assess linear models
know when you should move beyond linearity
Reading

- An Introduction to Statistical Learning by James, Witten, Hastie & Tibshirani
November 2019

model categories depending on data
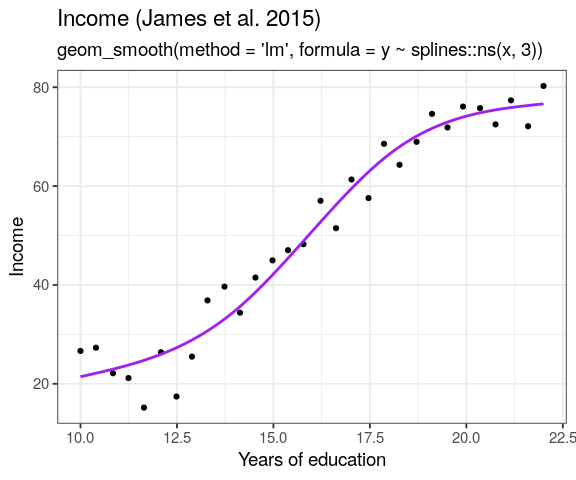
know when you should move beyond linearity

\[Y = f(X) + \epsilon\]
where:
\[\hat{Y} = \hat{f}(X)\]
Goal is to minimized: \[E(Y - \hat{Y})^2 = [f(X) - \hat{f}(X)]^2 + Var(\epsilon)\]
where \(Var(\epsilon)\) is the irreductible error.
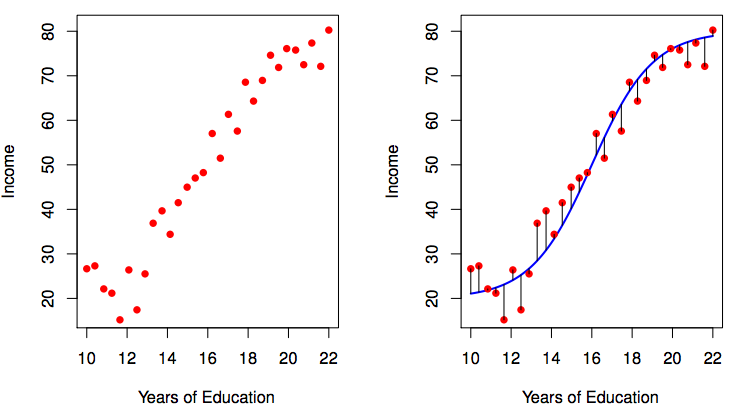
To estimate \(f\), one usually uses a limited number of observations.
this purple \(\hat{f}\) is different from the true blue \(f\)
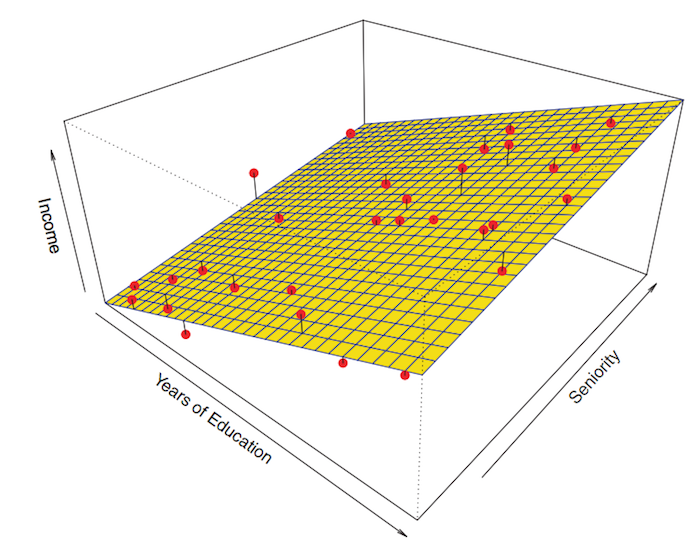
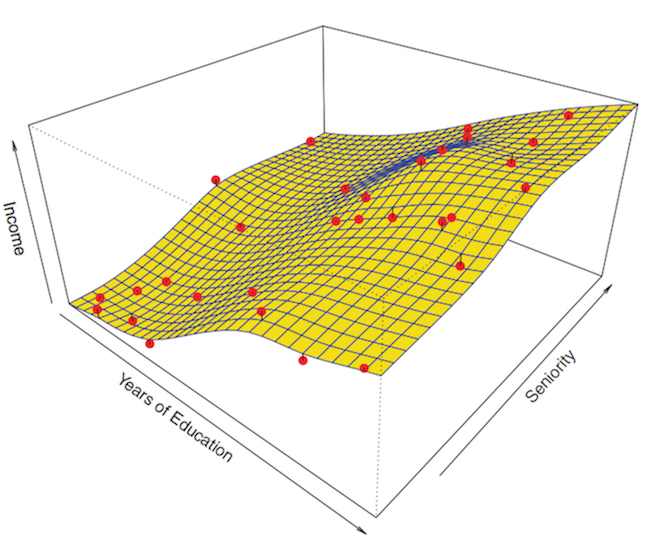
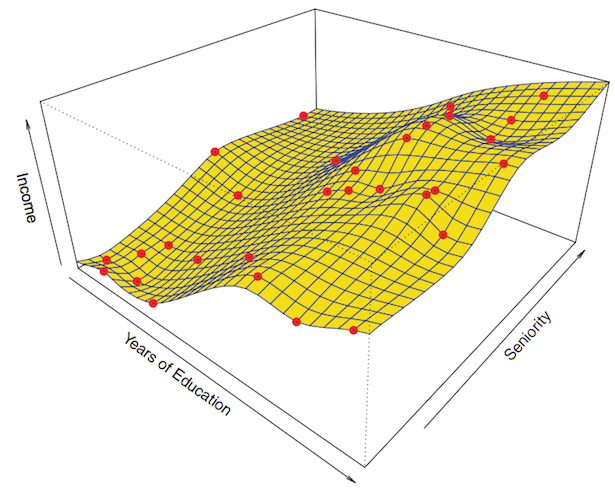
The first step of the parametric approach is done:
The general formula for p parameters gives p coefficients (slope) and \(\beta_0\) the intercept
\[f(X) = \beta_0 + \beta_1 \times X_1 + \beta_2 \times X_2 + \ldots + \beta_p \times X_p\]
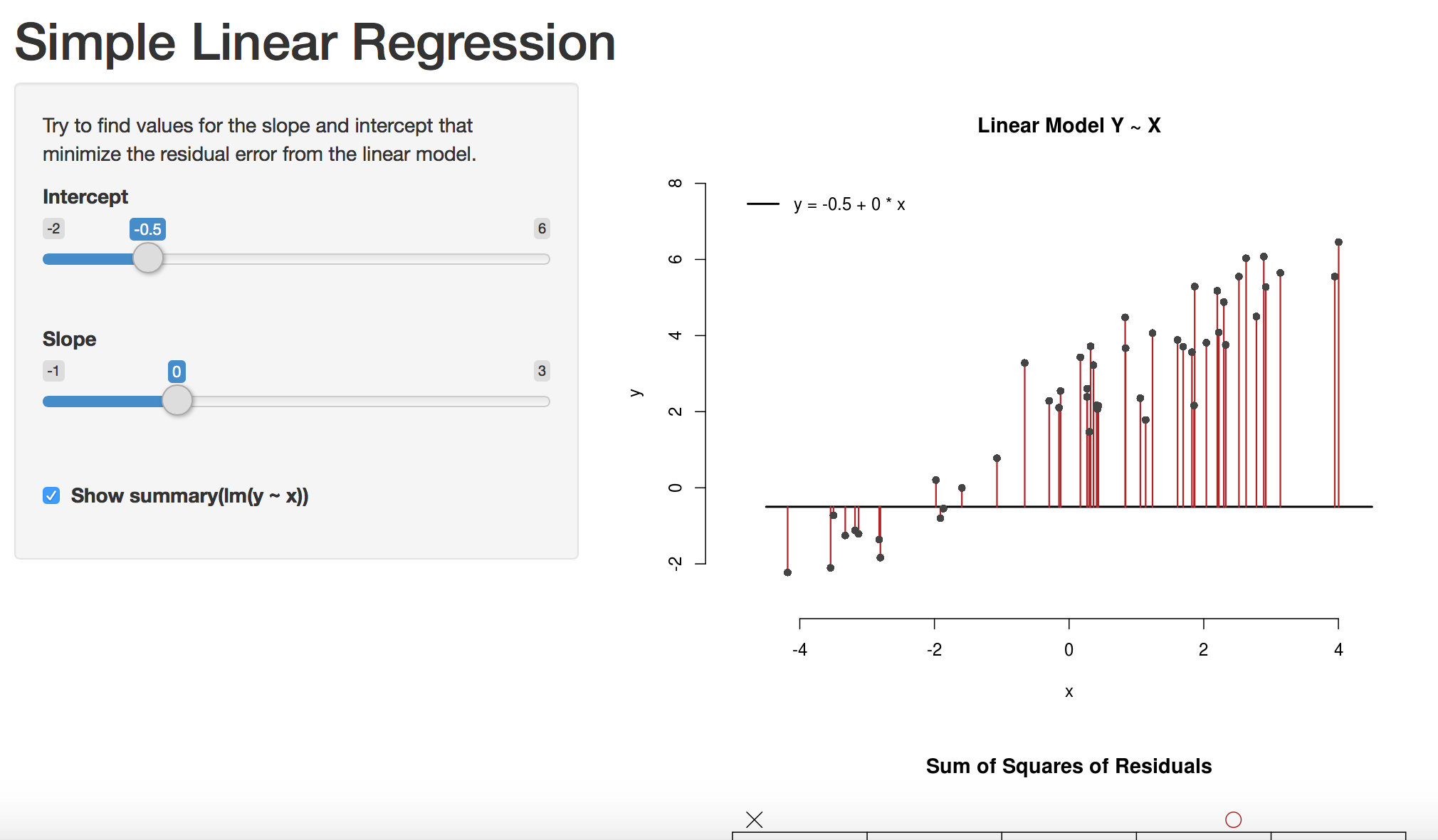
The model tries to minimize the reducible error, i. e the Sum of Squares of residuals
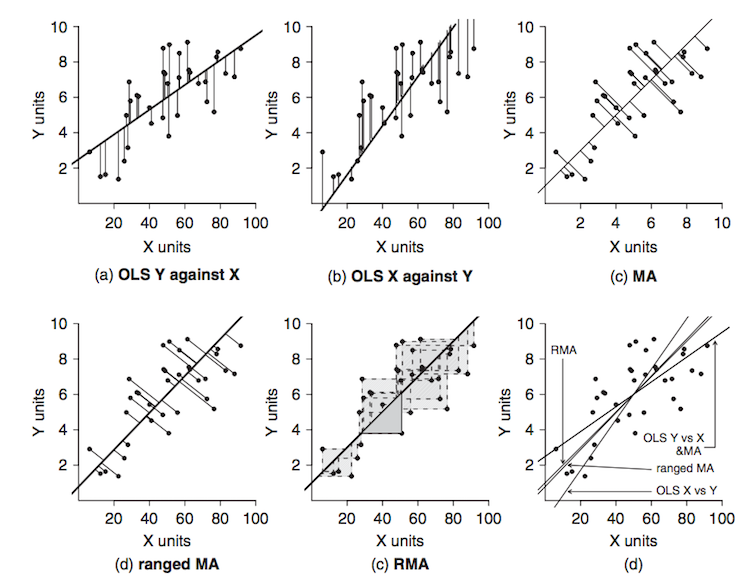
source: Logan M.
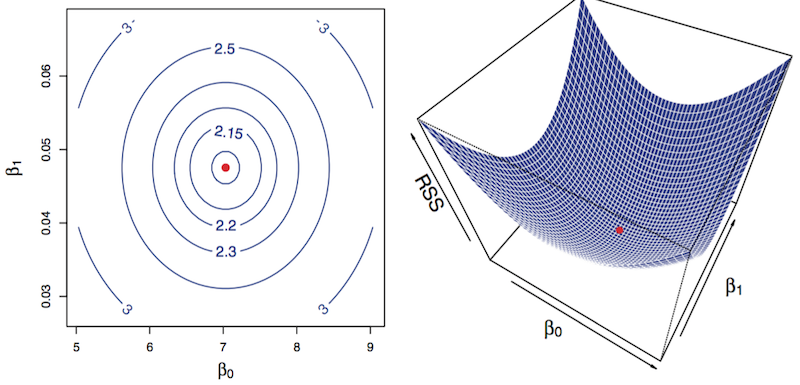
\[RSS = \sum_{i=1}^n{(y_i - \hat{y_i})^2}\]
estimate the intercept and the slope, \(\Rightarrow\) is the response increase for 1 unit of the predictor.
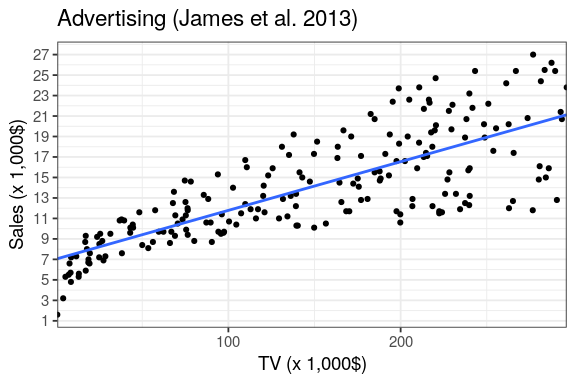
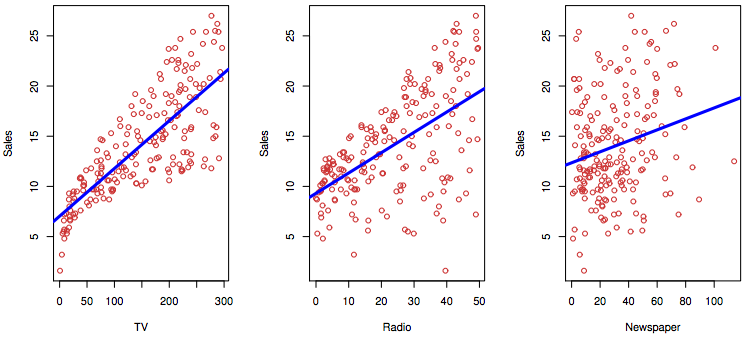
read_csv("data/Advertising.csv",
col_types = cols()) %>%
lm(Sales ~ TV, data = .) %>%
summary()
Call:
lm(formula = Sales ~ TV, data = .)
Residuals:
Min 1Q Median 3Q Max
-8.3860 -1.9545 -0.1913 2.0671 7.2124
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 7.032594 0.457843 15.36 <2e-16 ***
TV 0.047537 0.002691 17.67 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.259 on 198 degrees of freedom
Multiple R-squared: 0.6119, Adjusted R-squared: 0.6099
F-statistic: 312.1 on 1 and 198 DF, p-value: < 2.2e-16\[RSE = \sqrt{\frac{RSS}{n - 2}}\] why \(-2\) ?
\[SE(\hat{\beta_{1}})^2 = \frac{RSE}{\sum_{i=1}^n(x_i - \hat{x})^2}\]
with this standard error, one per coefficient, we can compute confidence intervals
read_csv("data/Advertising.csv") %>%
lm(Sales ~ TV, data = .) %>%
# tidy linear model using broom::tidy() into tibbles
tidy() %>%
mutate(CI95 = std.error * qt(0.975, df = (200 - 2)),
CI.L = estimate - CI95,
CI.R = estimate + CI95)
Parsed with column specification: cols( obs = col_double(), TV = col_double(), Radio = col_double(), Newspaper = col_double(), Sales = col_double() )
# A tibble: 2 x 8 term estimate std.error statistic p.value CI95 CI.L CI.R <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 (Intercept) 7.03 0.458 15.4 1.41e-35 0.903 6.13 7.94 2 TV 0.0475 0.00269 17.7 1.47e-42 0.00531 0.0422 0.0528
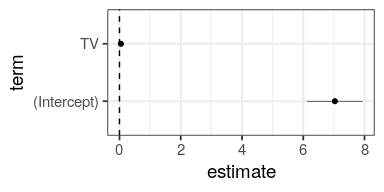
and the t-statistic for each coefficient \[t = \frac{\hat{\beta_1} - 0}{SE(\hat{\beta_1})}\]
RSE is a measure of the lack of fit
But, depends on sample size (units of Y)
\[R^2 = 1- \frac{RSS}{\sum(y_i - \bar{y})^2} = 1- \frac{RSS}{TSS}\]
the correlation coefficient, \(r^2\), is identical to the \(R^2\) and so the p values
Both regression and correlation examine the strength and signifiance of a relationship, regression analysis also explores the functional nature of the relationship Murray Logan
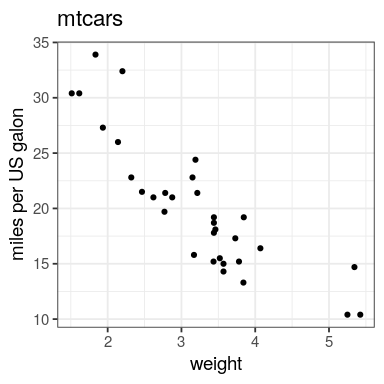
cor(mtcars$mpg, mtcars$wt)
[1] -0.8676594
cor(mtcars$mpg, mtcars$wt, method = "spearman")
[1] -0.886422
cor(mtcars$mpg, mtcars$wt, method = "kendall")
[1] -0.7278321
source: Logan M.
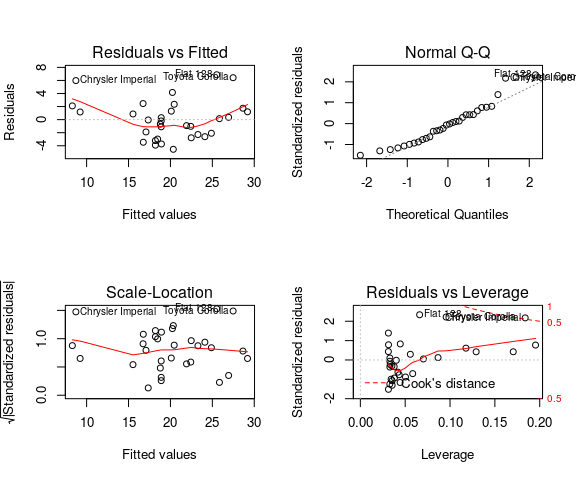
par(mfrow = (c(2, 2))) plot(lm(mpg ~ wt, data = mtcars))
library(ggfortify)
autoplot(lm(mpg ~ wt,
data = mtcars))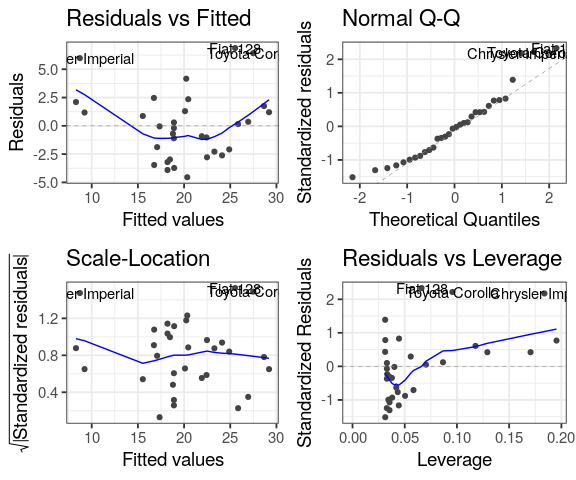
ggplot(mtcars, aes(x = wt, y = mpg)) + geom_point() + geom_smooth(method = "lm", se = FALSE)
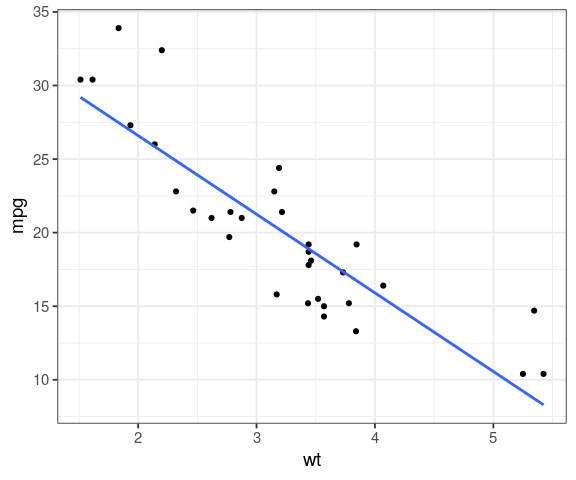
what about a car that weight 4,500 lb (~ 2 tons) ?
fit <- lm(mpg ~ wt, data = mtcars) # create linear model
Use the function coef to extract parameters slope and intercept
ggplot(mtcars, aes(x = wt, y = mpg)) +
geom_point() +
geom_smooth(method = "lm",
se = FALSE) +
# new unknown car
geom_vline(xintercept = 4.5) +
geom_hline(yintercept =
coef(fit)[1] + coef(fit)[2] * 4.5,
linetype = "dashed")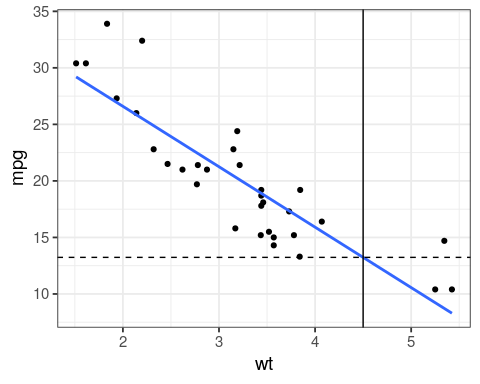
coef(fit)[1] + coef(fit)[2] * 4.5
(Intercept)
13.235 predict(fit, data.frame(wt = c(3, 4.5)))
1 2 21.25171 13.23500
geom_abline()ggplot(mtcars, aes(x = wt, y = mpg)) + geom_point() + geom_abline(slope = coef(fit)[2], intercept = coef(fit)[1]) + geom_smooth(method = "lm", se = FALSE, linetype = "dashed")
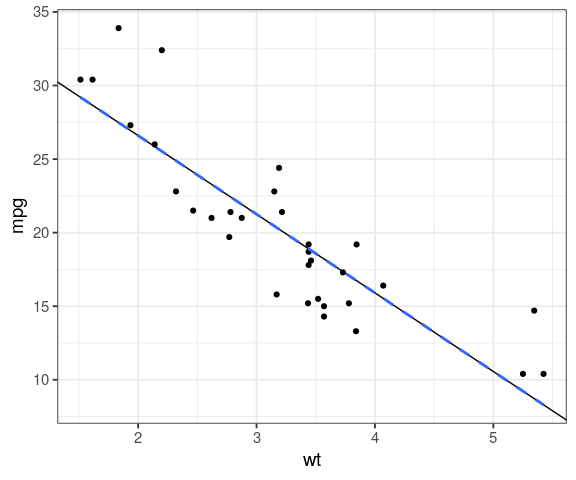
augmentbroom functionstidy(fit), coefficient estimates and hypotheses testingglance(fit), summary of the modelaugmenthead(augment(fit), 20)
# A tibble: 20 x 10 .rownames mpg wt .fitted .se.fit .resid .hat .sigma .cooksd <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 Mazda RX4 21 2.62 23.3 0.634 -2.28 0.0433 3.07 1.33e-2 2 Mazda RX… 21 2.88 21.9 0.571 -0.920 0.0352 3.09 1.72e-3 3 Datsun 7… 22.8 2.32 24.9 0.736 -2.09 0.0584 3.07 1.54e-2 4 Hornet 4… 21.4 3.22 20.1 0.538 1.30 0.0313 3.09 3.02e-3 5 Hornet S… 18.7 3.44 18.9 0.553 -0.200 0.0329 3.10 7.60e-5 6 Valiant 18.1 3.46 18.8 0.555 -0.693 0.0332 3.10 9.21e-4 7 Duster 3… 14.3 3.57 18.2 0.573 -3.91 0.0354 3.01 3.13e-2 8 Merc 240D 24.4 3.19 20.2 0.539 4.16 0.0313 3.00 3.11e-2 9 Merc 230 22.8 3.15 20.5 0.540 2.35 0.0314 3.07 9.96e-3 10 Merc 280 19.2 3.44 18.9 0.553 0.300 0.0329 3.10 1.71e-4 11 Merc 280C 17.8 3.44 18.9 0.553 -1.10 0.0329 3.09 2.30e-3 12 Merc 450… 16.4 4.07 15.5 0.719 0.867 0.0558 3.09 2.53e-3 13 Merc 450… 17.3 3.73 17.4 0.610 -0.0502 0.0401 3.10 5.92e-6 14 Merc 450… 15.2 3.78 17.1 0.624 -1.88 0.0419 3.08 8.73e-3 15 Cadillac… 10.4 5.25 9.23 1.26 1.17 0.170 3.09 1.84e-2 16 Lincoln … 10.4 5.42 8.30 1.35 2.10 0.195 3.07 7.19e-2 17 Chrysler… 14.7 5.34 8.72 1.31 5.98 0.184 2.84 5.32e-1 18 Fiat 128 32.4 2.2 25.5 0.783 6.87 0.0661 2.80 1.93e-1 19 Honda Ci… 30.4 1.62 28.7 1.05 1.75 0.118 3.08 2.49e-2 20 Toyota C… 33.9 1.84 27.5 0.942 6.42 0.0956 2.83 2.60e-1 # … with 1 more variable: .std.resid <dbl>
# create linear model fit <- lm(mpg ~ wt, data = mtcars) # try this command: predict(fit)[1:10]
predict call the predicted fuel consumption for the original car weights that were used to built the model.using predict without argumentspredict(fit)
Mazda RX4 Mazda RX4 Wag Datsun 710
23.282611 21.919770 24.885952
Hornet 4 Drive Hornet Sportabout Valiant
20.102650 18.900144 18.793255
Duster 360 Merc 240D Merc 230
18.205363 20.236262 20.450041
Merc 280 Merc 280C Merc 450SE
18.900144 18.900144 15.533127
Merc 450SL Merc 450SLC Cadillac Fleetwood
17.350247 17.083024 9.226650
Lincoln Continental Chrysler Imperial Fiat 128
8.296712 8.718926 25.527289
Honda Civic Toyota Corolla Toyota Corona
28.653805 27.478021 24.111004
Dodge Challenger AMC Javelin Camaro Z28
18.472586 18.926866 16.762355
Pontiac Firebird Fiat X1-9 Porsche 914-2
16.735633 26.943574 25.847957
Lotus Europa Ford Pantera L Ferrari Dino
29.198941 20.343151 22.480940
Maserati Bora Volvo 142E
18.205363 22.427495 \[residuals = original - predicted\]
my_residuals <- mtcars$mpg - predict(fit) head(my_residuals)
Mazda RX4 Mazda RX4 Wag Datsun 710 Hornet 4 Drive
-2.2826106 -0.9197704 -2.0859521 1.2973499
Hornet Sportabout Valiant
-0.2001440 -0.6932545 R has a residuals functionthat does the job
head(residuals(fit))
Mazda RX4 Mazda RX4 Wag Datsun 710 Hornet 4 Drive
-2.2826106 -0.9197704 -2.0859521 1.2973499
Hornet Sportabout Valiant
-0.2001440 -0.6932545 See also summary(residuals(fit)) that shows the upper part of summary(fit)
\[RSS = \sum_{i=1}^n{(y_i - \hat{y_i})^2}\]
sum(residuals(fit)^2)
[1] 278.3219
library(broom) glance(fit)
# A tibble: 1 x 11
r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
<dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.753 0.745 3.05 91.4 1.29e-10 2 -80.0 166. 170.
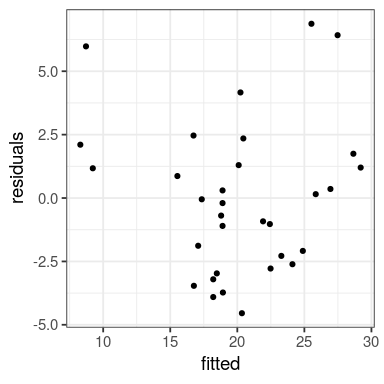
# … with 2 more variables: deviance <dbl>, df.residual <int>tibble(fitted = predict(fit),
residuals = residuals(fit)) %>%
ggplot() +
geom_point(aes(fitted, residuals))
tibble(x = residuals(fit)) %>% ggplot() + stat_qq(aes(sample = x))
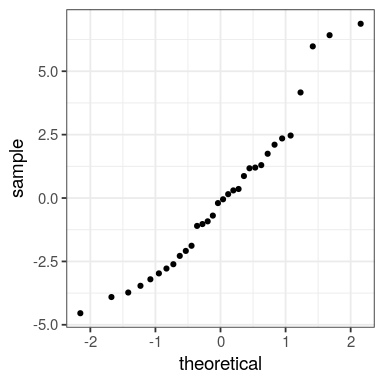
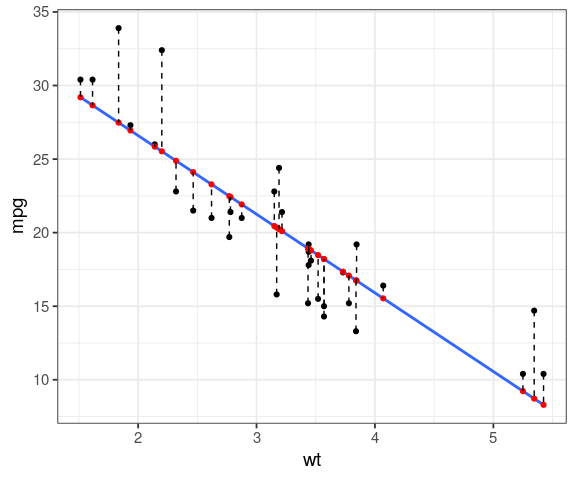
We want the predicted values in red, residuals in dashed black
Minimizing the error sum of squares does not resolve everything.
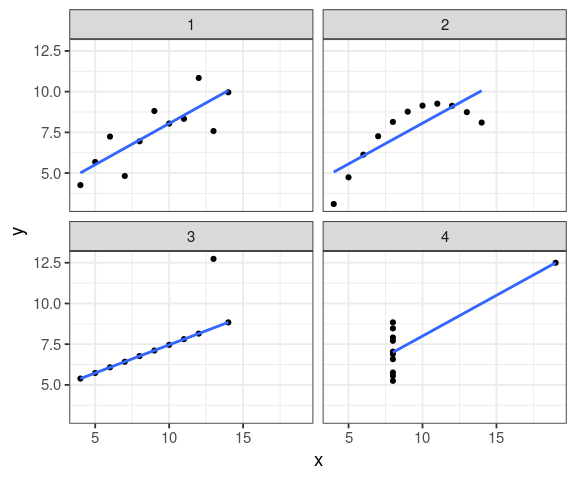
anscombe
x1 x2 x3 x4 y1 y2 y3 y4 1 10 10 10 8 8.04 9.14 7.46 6.58 2 8 8 8 8 6.95 8.14 6.77 5.76 3 13 13 13 8 7.58 8.74 12.74 7.71 4 9 9 9 8 8.81 8.77 7.11 8.84 5 11 11 11 8 8.33 9.26 7.81 8.47 6 14 14 14 8 9.96 8.10 8.84 7.04 7 6 6 6 8 7.24 6.13 6.08 5.25 8 4 4 4 19 4.26 3.10 5.39 12.50 9 12 12 12 8 10.84 9.13 8.15 5.56 10 7 7 7 8 4.82 7.26 6.42 7.91 11 5 5 5 8 5.68 4.74 5.73 6.89
Is this data.frame tidy?
anscombe_tidy <- anscombe %>%
pivot_longer(
everything(),
# .value is a placeholder that
# will received x and y
names_to = c(".value", "set"),
# regular expression to capture
names_pattern = "(.)(.)"
) %>%
as_tibble()
anscombe_tidy
# A tibble: 44 x 3 set x y <chr> <dbl> <dbl> 1 1 10 8.04 2 2 10 9.14 3 3 10 7.46 4 4 8 6.58 5 1 8 6.95 6 2 8 8.14 7 3 8 6.77 8 4 8 5.76 9 1 13 7.58 10 2 13 8.74 # … with 34 more rows
broom::tidy to get a tibble backanscombe_tidy %>%
group_nest(set) %>%
mutate(model = map(data, ~ lm(y ~ x, data = .x)),
tidy = map(model, tidy)) %>%
unnest(tidy, .drop = TRUE)
Warning: The `.drop` argument of `unnest()` is deprecated as of tidyr 1.0.0. All list-columns are now preserved. This warning is displayed once per session. Call `lifecycle::last_warnings()` to see where this warning was generated.
# A tibble: 8 x 8 set data model term estimate std.error statistic p.value <chr> <list> <lis> <chr> <dbl> <dbl> <dbl> <dbl> 1 1 <tibble [11 ×… <lm> (Interce… 3.00 1.12 2.67 0.0257 2 1 <tibble [11 ×… <lm> x 0.500 0.118 4.24 0.00217 3 2 <tibble [11 ×… <lm> (Interce… 3.00 1.13 2.67 0.0258 4 2 <tibble [11 ×… <lm> x 0.5 0.118 4.24 0.00218 5 3 <tibble [11 ×… <lm> (Interce… 3.00 1.12 2.67 0.0256 6 3 <tibble [11 ×… <lm> x 0.500 0.118 4.24 0.00218 7 4 <tibble [11 ×… <lm> (Interce… 3.00 1.12 2.67 0.0256 8 4 <tibble [11 ×… <lm> x 0.500 0.118 4.24 0.00216
compare the \(R^2\) using glance
anscombe_tidy %>%
group_nest(set) %>%
mutate(model = map(data, ~ lm(y ~ x, data = .x)),
glance = map(model, glance)) %>%
unnest(glance, .drop = TRUE)
# A tibble: 4 x 14 set data model r.squared adj.r.squared sigma statistic p.value df <chr> <lis> <lis> <dbl> <dbl> <dbl> <dbl> <dbl> <int> 1 1 <tib… <lm> 0.667 0.629 1.24 18.0 0.00217 2 2 2 <tib… <lm> 0.666 0.629 1.24 18.0 0.00218 2 3 3 <tib… <lm> 0.666 0.629 1.24 18.0 0.00218 2 4 4 <tib… <lm> 0.667 0.630 1.24 18.0 0.00216 2 # … with 5 more variables: logLik <dbl>, AIC <dbl>, BIC <dbl>, # deviance <dbl>, df.residual <int>
all \(R^2\) are identical!
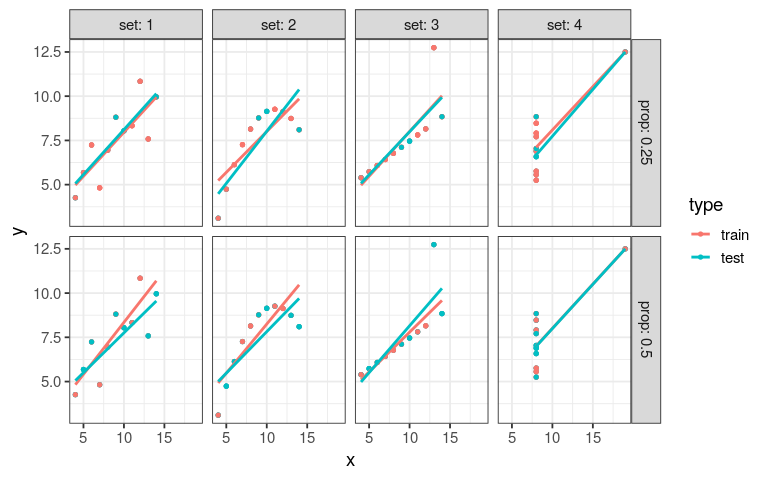
ggplot(anscombe_tidy, aes(x, y)) + geom_point() + facet_wrap(~ set, ncol = 2) + geom_smooth(method = "lm", se = FALSE)
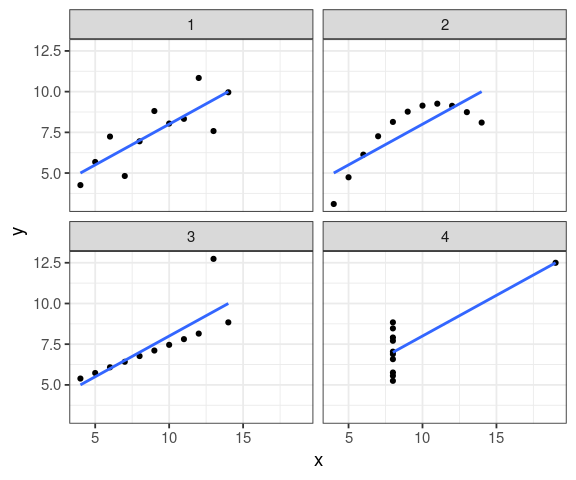
Luc Buissière has a nice way of saying it, from his former teacher Prof. Glenn Morris Univ. of Toronto:

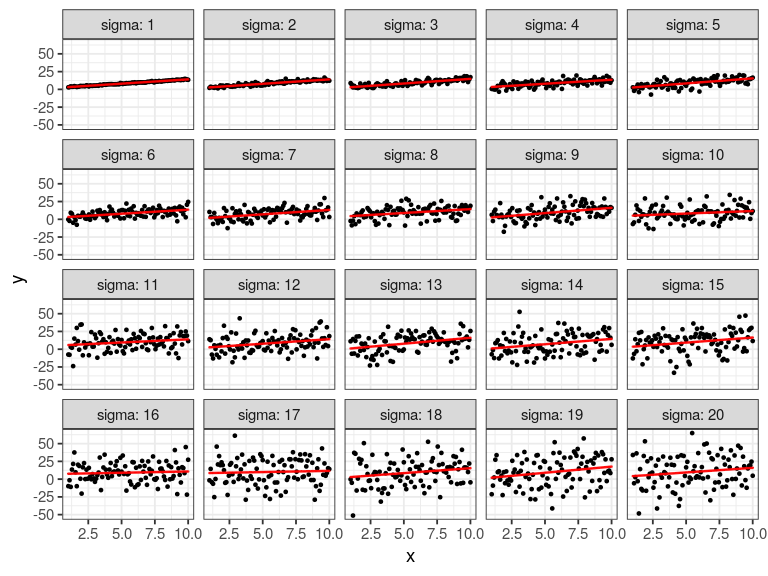
set.seed(123)
x <- seq(1, 10, length.out = 100) # our predictor
# our response; a function of x plus some random noise
map_dfr(seq(0.5, 20, length.out = 20),
~ lm(2 + 1.2 * x + rnorm(100, 0, sd = .x) ~ x) %>%
broom::glance()) %>%
mutate(sigma = seq(0.5, 20, length.out = 20)) %>%
ggplot(aes(x = sigma, y = r.squared)) +
geom_point(size = 2) +
geom_line()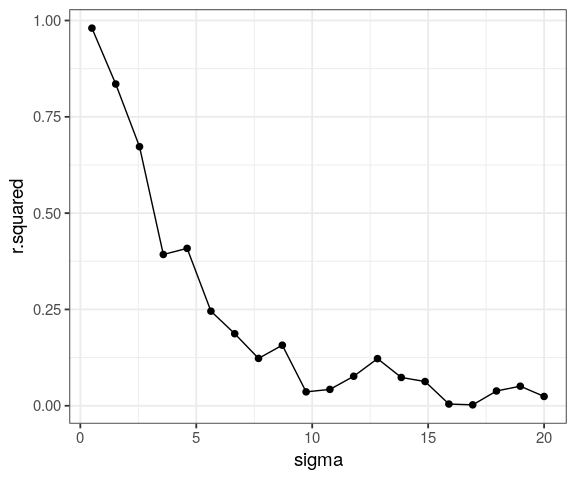
rlm (M-estimator Huber) weigths the outliersMASSlibrary(MASS)
anscombe_tidy %>%
group_nest(set) %>%
mutate(model = map(data,
~ rlm(y ~ x,
data = .x)),
tidy = map(model, tidy)) %>%
unnest(tidy)# A tibble: 8 x 7 set data model term estimate std.error statistic <chr> <list> <list> <chr> <dbl> <dbl> <dbl> 1 1 <tibble [11 × 2]> <rlm> (Intercept) 2.98 1.45 2.06 2 1 <tibble [11 × 2]> <rlm> x 0.506 0.152 3.34 3 2 <tibble [11 × 2]> <rlm> (Intercept) 3.06 1.31 2.33 4 2 <tibble [11 × 2]> <rlm> x 0.5 0.137 3.64 5 3 <tibble [11 × 2]> <rlm> (Intercept) 4.00 0.00404 990. 6 3 <tibble [11 × 2]> <rlm> x 0.346 0.000424 816. 7 4 <tibble [11 × 2]> <rlm> (Intercept) 3.00 1.26 2.38 8 4 <tibble [11 × 2]> <rlm> x 0.500 0.132 3.80
ggplot(anscombe_tidy, aes(x, y)) + geom_point() + facet_wrap(~ set, ncol = 2) + geom_smooth(method = "rlm", se = FALSE)
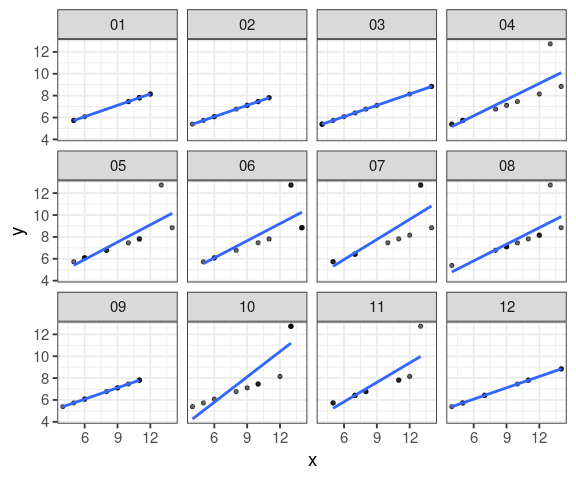
boot_set <- function(tib, dataset, rep = 1000) {
# per set using tidyeval
filter(tib, set == {{dataset}}) %>%
# generate 1000 dataset randomly
modelr::bootstrap(rep) %>%
mutate(model = map(strap, ~ lm(y ~ x, .x)),
tidy = map(model, tidy)) %>%
unnest(tidy)
}
#boot_set(anscombe_tidy, "I", 6)
my_sets <- set_names(1:4, nm = c("I", "II", "III", "IV"))
lm_boot <- map_dfr(my_sets, ~ boot_set(anscombe_tidy, .x), .id = "set")filter(anscombe_tidy, set == 3) %>% modelr::bootstrap(12) %>% mutate(boot = map(strap, as_tibble)) %>% unnest(boot) %>% ggplot(aes(x = x, y = y)) + geom_point(alpha = 0.6, size = 1.2) + geom_smooth(method = "lm", se = FALSE) + facet_wrap(~ .id)
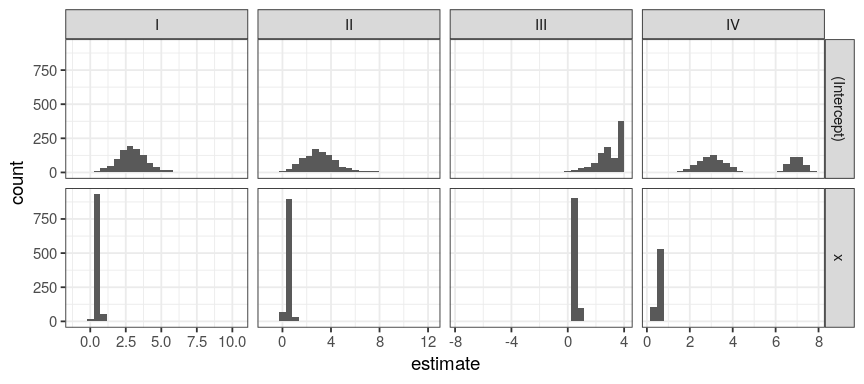
only I is displaying a normal distribution of its coefficients
crossing(set = 1:4,
prop = c(0.25, 0.5)) %>%
mutate(cross = map2(set, prop,
~ crossv_mset(tib = anscombe_tidy,
dataset = .x,
prop = .y)),
MSE_slope = map_dbl(cross, "MSE"),
MSE_inter = map_dbl(cross, "MSEIntercept")) %>%
pivot_longer(cols = starts_with("MSE"),
names_to = "term",
values_to = "MSE") %>%
ggplot(aes(x = set, y = MSE)) +
geom_col(aes(fill = factor(prop)),
position = "dodge") +
facet_wrap(~ term, ncol = 1) +
theme(legend.position = "bottom") +
labs(x = NULL, fill = "prop of test/train",
title = "Anscombe",
subtitle = "100 samples per data split")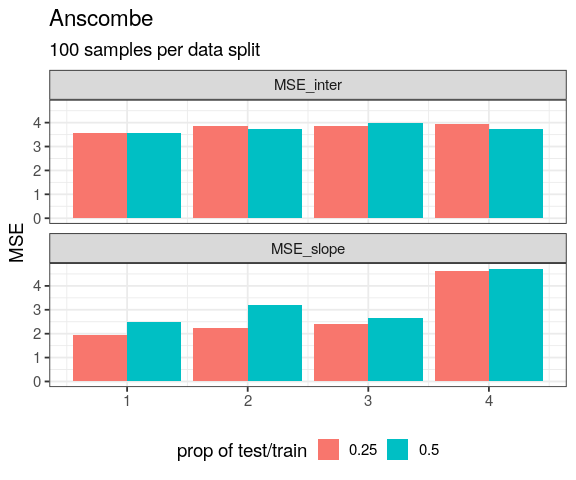
ref: David Robinson
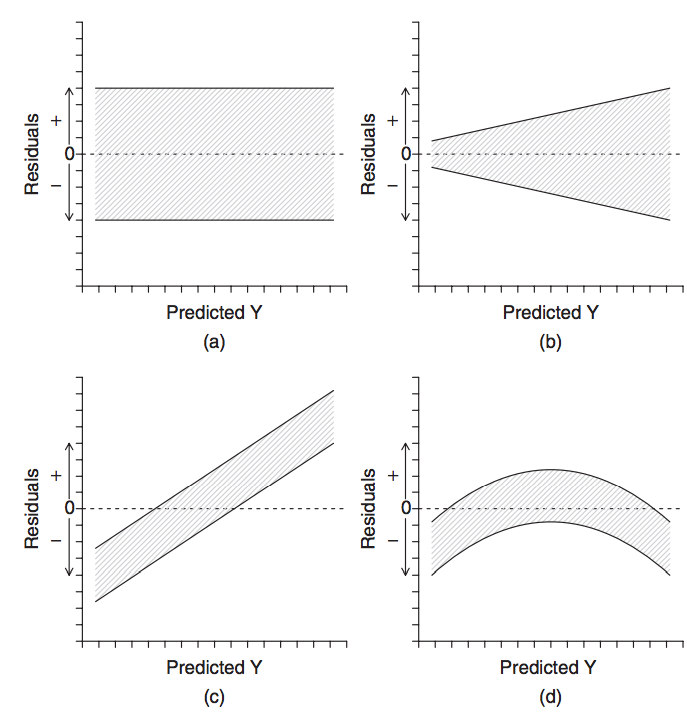
when trend is obviously non-linear, as seen in the residuals
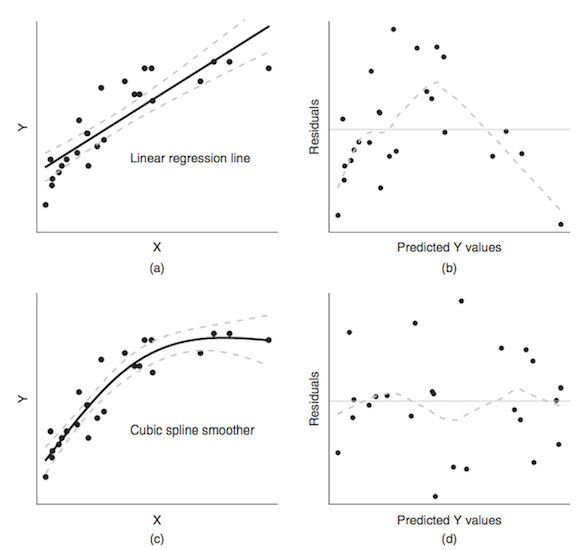
source: Logan M.
Some of the figures in this presentation are taken from:
An Introduction to Statistical Learning, with applications in R (Springer, 2013) with permission from the authors: G. James, D. Witten, T. Hastie and R. Tibshirani
Biostatistic Design and Analysis Using R. (Wiley-Blackwell, 2011) Murray Logan