You will learn to:

- basics of statistic
- term definition
- theoritical distributions
- compare 2 samples
October 2019




Clearly identify response variables from explanatory (or predictor) variables.
geom_smooth() with linear modelling
ggplot(mtcars, aes(x = wt, y = mpg)) + geom_point() + geom_smooth(method = "lm")
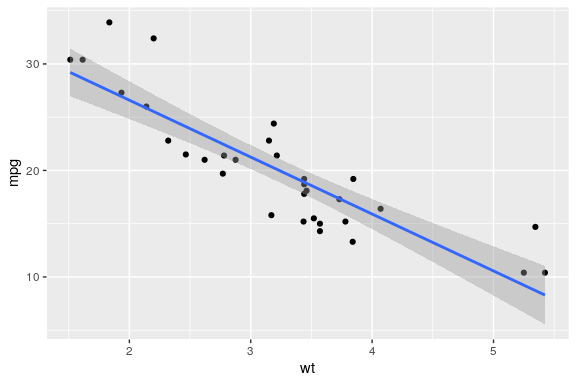
mpg) is the responsewt) is a predictorcor.test(mtcars$mpg, mtcars$wt)
Pearson's product-moment correlation
data: mtcars$mpg and mtcars$wt
t = -9.559, df = 30, p-value = 1.294e-10
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.9338264 -0.7440872
sample estimates:
cor
-0.8676594 cor.test(mtcars$wt, mtcars$mpg)
Pearson's product-moment correlation
data: mtcars$wt and mtcars$mpg
t = -9.559, df = 30, p-value = 1.294e-10
alternative hypothesis: true correlation is not equal to 0
95 percent confidence interval:
-0.9338264 -0.7440872
sample estimates:
cor
-0.8676594 summary(lm(mtcars$mpg ~ mtcars$wt))
Call:
lm(formula = mtcars$mpg ~ mtcars$wt)
Residuals:
Min 1Q Median 3Q Max
-4.5432 -2.3647 -0.1252 1.4096 6.8727
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 37.2851 1.8776 19.858 < 2e-16 ***
mtcars$wt -5.3445 0.5591 -9.559 1.29e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.046 on 30 degrees of freedom
Multiple R-squared: 0.7528, Adjusted R-squared: 0.7446
F-statistic: 91.38 on 1 and 30 DF, p-value: 1.294e-10cor(mtcars$wt, mtcars$mpg)
[1] -0.8676594
cor(mtcars$wt, mtcars$mpg)^2
[1] 0.7528328
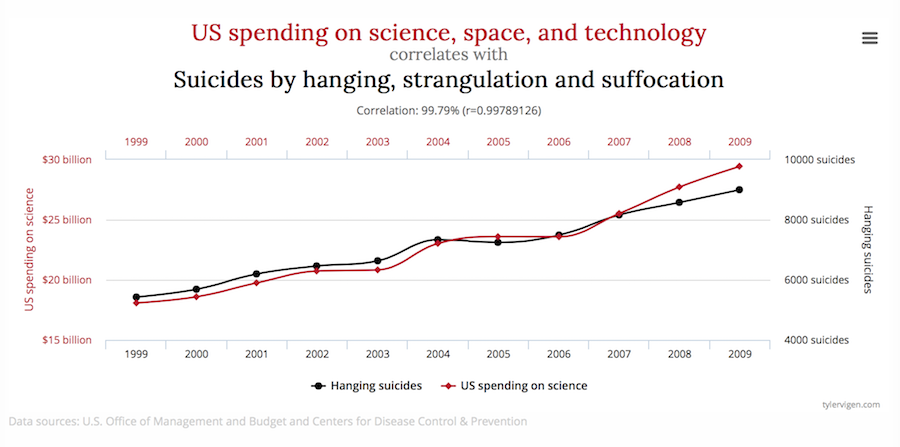
source: spurious correlations
Today’s Logic Lesson : Correlation v. Causation pic.twitter.com/ZJaLACw8tB
— Philosophy Matters (@PhilosophyMttrs) 30 septembre 2018
women$height[1:10]
[1] 58 59 60 61 62 63 64 65 66 67
ToothGrowth$supp[1:10]
[1] VC VC VC VC VC VC VC VC VC VC Levels: OJ VC
str(ToothGrowth$supp[1:10])
Factor w/ 2 levels "OJ","VC": 2 2 2 2 2 2 2 2 2 2
mtcars$cyl[1:10]
[1] 6 6 4 6 8 6 8 4 4 6
factor(mtcars$cyl[1:10])
[1] 6 6 4 6 8 6 8 4 4 6 Levels: 4 6 8
as.character(ToothGrowth$supp[1:10])
[1] "VC" "VC" "VC" "VC" "VC" "VC" "VC" "VC" "VC" "VC"
| Explanatory | Response | Method |
|---|---|---|
| continuous | continuous | regression |
| continuous | continuous | correlation |
| categorical | continuous | analysis of variance (anova) |
| continuous / categorical | continuous | analysis of covariance (ancova) |
| continuous | proportion | logistic regression |
| continuous | count | logit regression |
| continuous | time-at-death | survival analysis |
A good hypothesis is a falsifiable hypothesis Karl Popper
Which of these two is a good hypothesis?
p values are probabilities (\(0 \leq p \leq 1\)) that a particular result occurred by chance if \(H_0\) is true.
if \(H_0\) is true is almost never true. Instead a mixture of null and alternative hypotheses
p > 0.05 tells you the experiment is strictly underpowered and shouldn't be interpreted at all.
— John Myles White (@johnmyleswhite) 26 janvier 2018
p < 0.001 tells you that you've found something worth running multiple studies on.
and are made at a certain threshold

Attaching package: 'kableExtra'
The following object is masked from 'package:dplyr':
group_rows
Warning: funs() is soft deprecated as of dplyr 0.8.0 Please use a list of either functions or lambdas: # Simple named list: list(mean = mean, median = median) # Auto named with `tibble::lst()`: tibble::lst(mean, median) # Using lambdas list(~ mean(., trim = .2), ~ median(., na.rm = TRUE)) This warning is displayed once per session.
| decision on H0 | True | False |
|---|---|---|
| Reject | Type I error | Correct |
| Accept | Correct | Type II error |
the power of a test is \(1 - \beta\)
crossing(n = 3:30,
delta = NULL,
sig.level = .05,
power = seq(.3, .9, .2),
sd = 1,
type = c("two.sample")) %>%
pmap(.l = ., .f = power.t.test) %>%
map_df(tidy) %>%
ggplot(aes(x = n, y = delta)) +
geom_line(aes(colour = percent(power)),
size = 1.1) +
geom_hline(yintercept = 2^0.5,
linetype = "dashed") +
annotate("text", x = 25, y = 1.6,
label = "log[2]*\" = 0.5\"",
parse = TRUE, size = 6) +
scale_y_continuous(labels = percent) +
scale_x_continuous(breaks = c(3,5,10,20,30)) +
theme_minimal(20) +
labs(x = "# observations (both samples)",
y = "true difference of means",
color = "power",
title = "Power of 2-samples t-test ",
subtitle = "for sd = 1")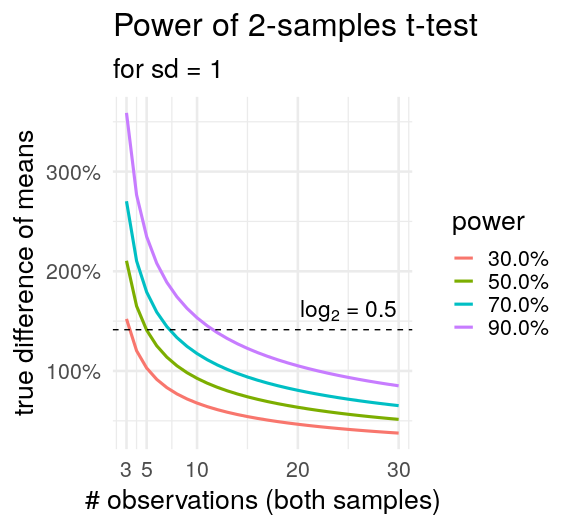
Your experimental design should take care of two key concepts
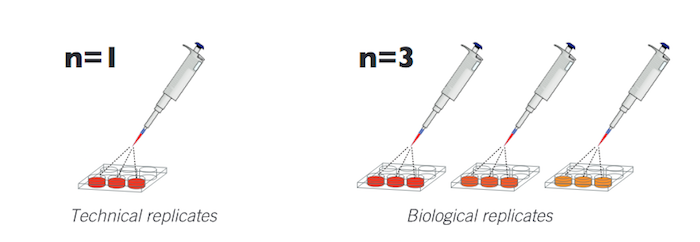
source asbmb.org
this implies that we have models, distributions that we know and can be used to assess our data.
 Two parameters
Two parameters
For smaller sample sizes, one more parameter: degrees of freedom. \[df = n - p\]
Thus, 3 parameters:
Larger incertainty leads to bigger tails
Let’s say we have 5 observations with a known average of 4.
| 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|
| 2 |
average = 2
| 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|
| 2 | 7 |
average = 4.5
| 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|
| 2 | 7 | 4 |
average = 4.3333333
| 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|
| 2 | 7 | 4 | 0 |
average = 3.25
and the average must be 4
we must have a sum of 20.
\[20 - (2 + 7 + 4 + 0) = 7\]
| 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|
| 2 | 7 | 4 | 0 | 7 |
average = 4
\[df = n - 1 = 5 - 1 = 4\]
suffers from extreme values
\[\mu = \frac{\sum x}{n}\]
middle value, sort values:
value that occurs the most frequently
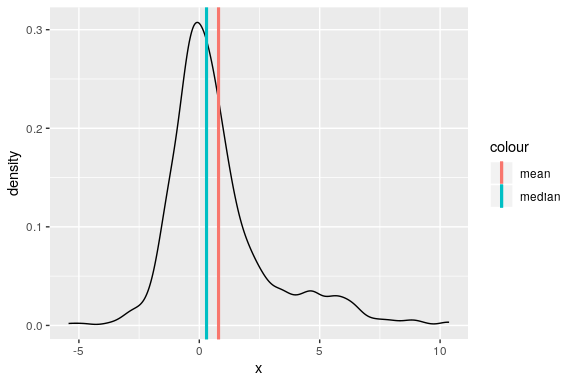
set.seed(123) dens <- data_frame(x = c(rnorm(500), rnorm(200, 3, 3)))
Warning: `data_frame()` is deprecated, use `tibble()`. This warning is displayed once per session.
ggplot(dens) + geom_line(aes(x), stat = "density") + geom_vline(aes(xintercept = mean(x), colour = "mean"), size = 1.1) + geom_vline(aes(xintercept = median(x), colour = "median"), size = 1.1) -> p p
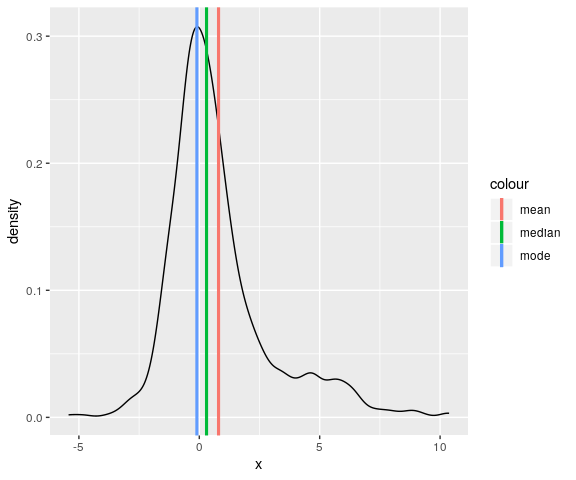
dens_mode <- density(dens$x)$x[which.max(density(dens$x)$y)] p + geom_vline(aes(xintercept = dens_mode, colour = "mode"), size = 1.1)
\[variance = \frac{sum\,of\,squares}{degrees\,of\,freedom}\]
\[residual = observed\,value - predicted\,value\]
x has a mean of \[3.0 \pm 0.365 (1 s.e, n = 10)\]
\[SE_\bar{y} = \sqrt{\frac{s^2}{n}}\]
A CI is how many t standard error to be expected for a given interval
\[confidence\,interval = t \times standard\,error\]
garden
# A tibble: 10 x 3
gardenA gardenB gardenC
<dbl> <dbl> <dbl>
1 3 5 3
2 4 5 3
3 4 6 2
4 3 7 1
5 2 4 10
6 3 4 4
7 1 3 3
8 3 5 11
9 5 6 3
10 2 5 10mean(garden$gardenA)
[1] 3
qt(0.025, df = 9)
[1] -2.262157
qt(0.975, df = 9)
[1] 2.262157
qt(0.975, df = 9) * sqrt(var(garden$gardenA) / length(garden$gardenA))
[1] 0.826023
\(\;\bar{x} = 3.0 \pm 0.83 (CI_{95\%})\)
mean() var() sd() # median absolute deviation mad()
a <- garden$gardenA mean(a)
[1] 3
var(a)
[1] 1.333333
sd(a)
[1] 1.154701
mad(a) # median absolute deviation
[1] 1.4826
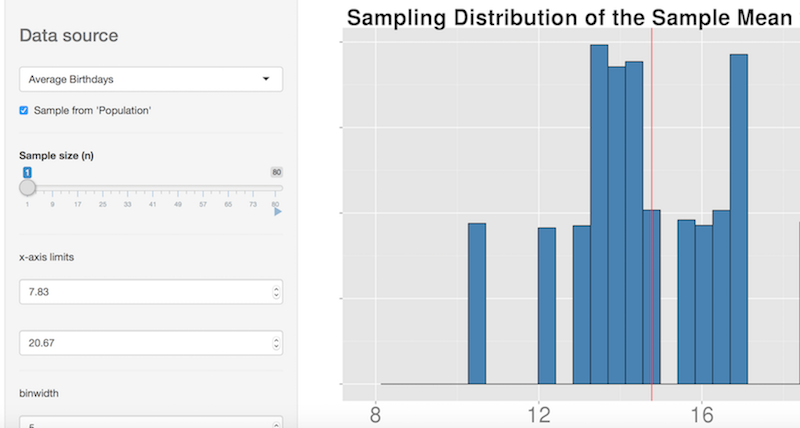
https://rpubs.com/nishantsbi/112044 by nishantsbi
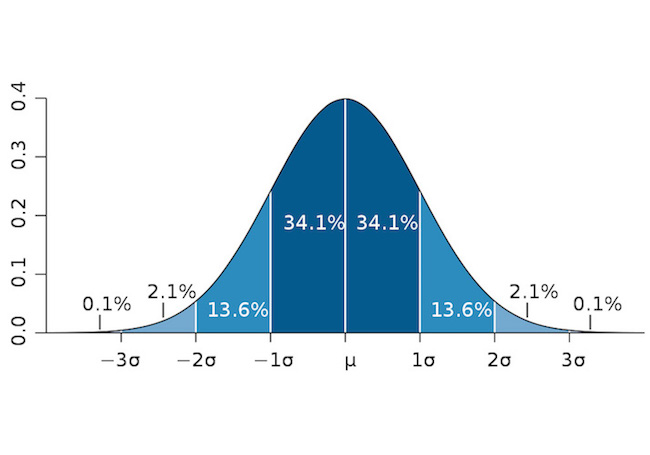
also called standardized scores, they refer to how many standard deviation to the mean.
\[z = \frac{x - \mu}{\sigma}\]
1 - 2 * pnorm(-2)
[1] 0.9544997
z <- qnorm(0.025) z
[1] -1.959964
1 - 2 * pnorm(z)
[1] 0.95
p, q, r for the normal distribution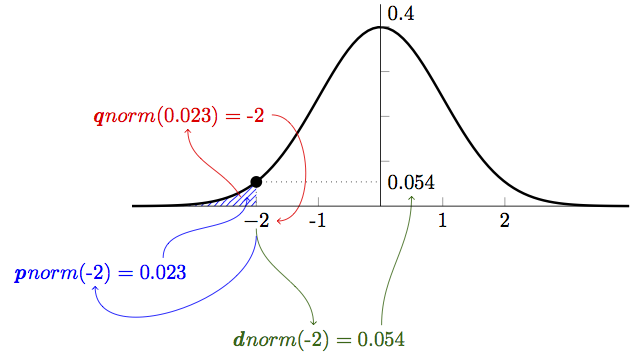
by Eric Koncina
Suppose we have measured the height of 100 people: The mean height is 170 cm and the standard deviation is 8 cm.
how much of the population is shorter than 160 cm?
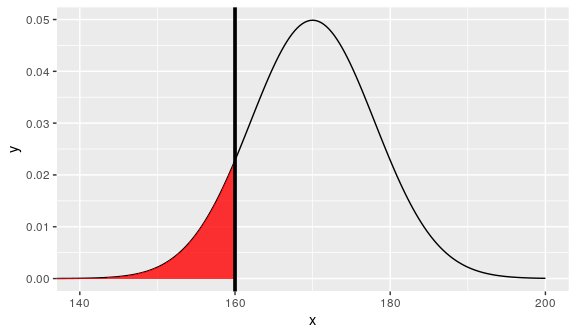
z <- (160 - 170) / 8 z
[1] -1.25
i.e area under the bell shape.
pnorm(z)
[1] 0.1056498
paste(round(pnorm(z) * 100, 2), "%")
[1] "10.56 %"
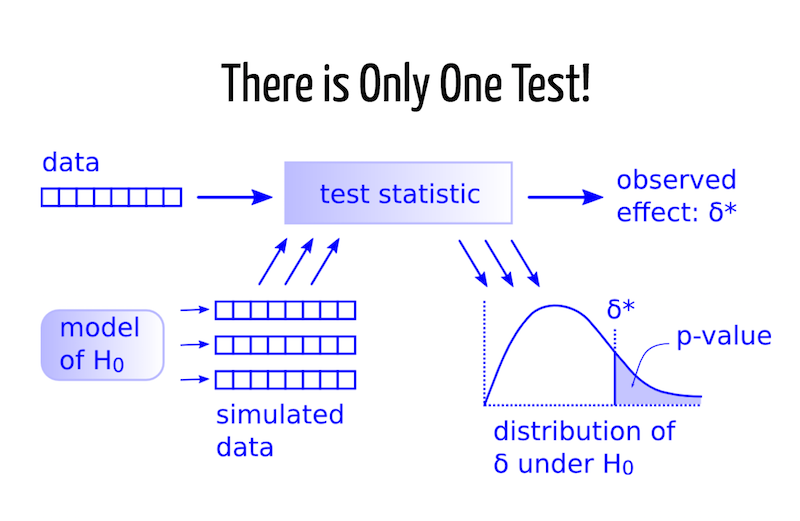
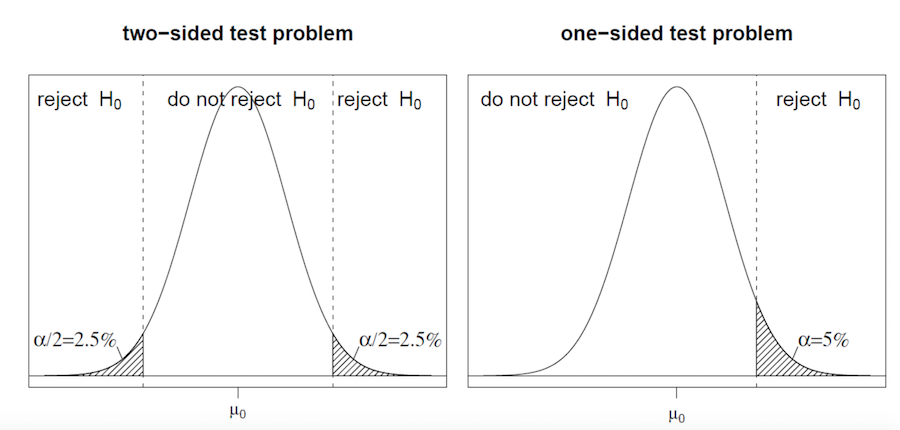
set.seed(123) rn <- rnorm(100) shapiro.test(rn)
Shapiro-Wilk normality test
data: rn
W = 0.99388, p-value = 0.9349t.test(garden$gardenA)
One Sample t-test
data: garden$gardenA
t = 8.2158, df = 9, p-value = 1.788e-05
alternative hypothesis: true mean is not equal to 0
95 percent confidence interval:
2.173977 3.826023
sample estimates:
mean of x
3 \[\mu\ != \bar{x}\]
mean(mtcars$mpg)
[1] 20.09062
t.test(mtcars$mpg, mu = 18, alternative = "two.sided")
One Sample t-test
data: mtcars$mpg
t = 1.9622, df = 31, p-value = 0.05876
alternative hypothesis: true mean is not equal to 18
95 percent confidence interval:
17.91768 22.26357
sample estimates:
mean of x
20.09062 var.test(sample1, sample2)
Divide the larger variance by the smaller one.
using the garden dataset.
var(garden$gardenA)
[1] 1.333333
var(garden$gardenC)
[1] 14.22222
F.ratio <- var(garden$gardenC) / var(garden$gardenA) F.ratio
[1] 10.66667
tibble(x = seq(-2, 10, length.out = 500),
y = df(x, 9, 9)) %>%
ggplot() +
geom_line(aes(x = x, y = y)) +
theme_bw(18)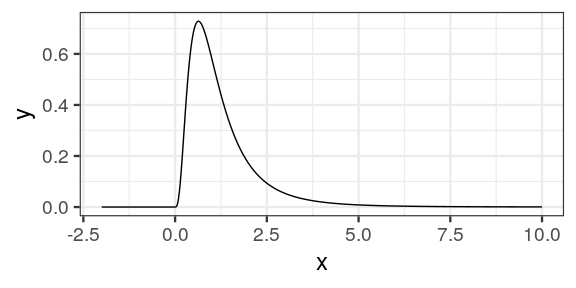
why no values < 0?
no values < 0, mind the degrees of freedom!
qf(0.975, 9, 9)
[1] 4.025994
Could you already accept or reject \(H_0\)?
2 * (1 - pf(F.ratio, 9, 9))
[1] 0.001624199
var.test(garden$gardenA, garden$gardenC)
F test to compare two variances
data: garden$gardenA and garden$gardenC
F = 0.09375, num df = 9, denom df = 9, p-value = 0.001624
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.02328617 0.37743695
sample estimates:
ratio of variances
0.09375 if not normal?
\[ t = \frac{difference\,between\,two\,means}{standard\,error\,of\,the\,difference} = \frac{ \overline{y_A} - \overline{y_B}}{s.e_{diff}}\]
the variance of their difference is the sum of their variances.
\[s.e_{diff} = \sqrt{\frac{s_A^2}{n_A} + \frac{s_B^2}{n_B}}\]
but let’s see for gardenA and B
var.test(garden$gardenA, garden$gardenB)
F test to compare two variances
data: garden$gardenA and garden$gardenB
F = 1, num df = 9, denom df = 9, p-value = 1
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.2483859 4.0259942
sample estimates:
ratio of variances
1 library(broom) tidy(shapiro.test(garden$gardenA))
# A tibble: 1 x 3
statistic p.value method
<dbl> <dbl> <chr>
1 0.953 0.703 Shapiro-Wilk normality testtidy(shapiro.test(garden$gardenB))
# A tibble: 1 x 3
statistic p.value method
<dbl> <dbl> <chr>
1 0.953 0.703 Shapiro-Wilk normality testsince two different gardens
compute it!
diff <- mean(garden$gardenA) - mean(garden$gardenB) diff
[1] -2
t <- diff / (sqrt((var(garden$gardenA) / 10) + (var(garden$gardenB) / 10))) t # sign does not matter, how many df?
[1] -3.872983
2 * pt(t, df = 18)
[1] 0.001114539
t.test(garden$gardenA, garden$gardenB,
var.equal = FALSE)
Welch Two Sample t-test
data: garden$gardenA and garden$gardenB
t = -3.873, df = 18, p-value = 0.001115
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-3.0849115 -0.9150885
sample estimates:
mean of x mean of y
3 5 t.test(garden$gardenA, garden$gardenB,
var.equal = TRUE)
Two Sample t-test
data: garden$gardenA and garden$gardenB
t = -3.873, df = 18, p-value = 0.001115
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-3.0849115 -0.9150885
sample estimates:
mean of x mean of y
3 5 t.test(x, y, paired = TRUE)
= a one-sample t test with the difference of samples
wilcox.test()
paired = FALSE, for independent samples, aka Mann-Whitney’s test.paired = TRUE, for dependent samples, aka Wilcoxon.garden %>% # go for long format gather(garden, ozone) %>% # focus on A and B filter(garden != "gardenC") %>% mutate(rank = rank(ozone, ties.method = "average")) %>% # do the grouping AFTER ranking group_by(garden) %>% # get the sum! summarise(sum(rank))
# A tibble: 2 x 2 garden `sum(rank)` <chr> <dbl> 1 gardenA 66 2 gardenB 144
value has to be < for both \(n = 10\) samples
qwilcox(0.975, 10, 10)
[1] 76
sum(rank).bind_rows(
tidy(wilcox.test(
garden$gardenA,
garden$gardenB)),
tidy(t.test(
garden$gardenA,
garden$gardenB)))# A tibble: 2 x 10
statistic p.value method alternative estimate estimate1 estimate2
<dbl> <dbl> <chr> <chr> <dbl> <dbl> <dbl>
1 11 0.00299 Wilco… two.sided NA NA NA
2 -3.87 0.00111 Welch… two.sided -2 3 5
# … with 3 more variables: parameter <dbl>, conf.low <dbl>,
# conf.high <dbl>non-parametric tests are more conservative with less power
for \(n=3\) power is zero. Minimum is \(n=4\)
ks.test()
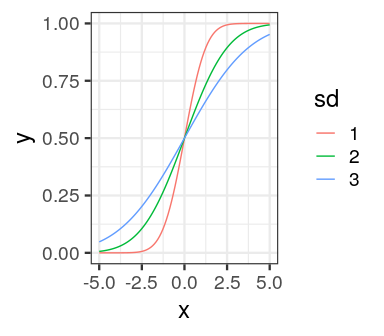
tibble(x = seq(-5, 5, length.out = 100),
`1` = pnorm(x),
`2` = pnorm(x, 0, 2),
`3` = pnorm(x, 0, 3)) %>%
gather(sd, y, -x) %>%
ggplot(aes(x, y, colour = sd)) +
geom_line() +
theme_bw(18)