You will learn to:

- (re)view some R base
- get the different data types:
numeric,logical,factor… - understand what is a
list, avector, adata.frame…
- no tidyverse, but remember it is built on base
October 2019

numeric, logical, factor …list, a vector, a data.frame …+: addition-: subtraction*: multiplication/: division^ or **: exponentiation%%: modulo (remainder after division)%/%: integer divisionR will:
If you need to change the priority during the evaluation, use parentheses – i.e. ( and ) – to group calculations.
9 / 2 # floating division
[1] 4.5
9 %/% 2 # integer division
[1] 4
9 %% 2 # remainder
[1] 1
(1:10 %/% 3) * 3 # int div
[1] 0 0 3 3 3 6 6 6 9 9
1:10 %% 3 # remainder
[1] 1 2 0 1 2 0 1 2 0 1
(1:10 %% 3) + (1:10 %/% 3) * 3 # sum up
[1] 1 2 3 4 5 6 7 8 9 10
We could let base down, but the tidyverse is wrapping around it. Some functions need to be known

I teach them X just to show them how much easier Y is
teaching programming is hard, don’t make it harder
When you start writing a loop then turn it into dplyr#rstats pic.twitter.com/M0gXUXuYCP
— David Robinson (@drob) 22 Feb 2016
| Type | Example |
|---|---|
| numeric | integer (2), double (2.34) |
| character (strings) | “tidyverse !” |
| boolean | TRUE / FALSE |
| complex | 2+0i |
NA # not available, missing data NA_real_ NA_integer_ NA_character_ NA_complex_ NULL # empty -Inf/Inf # infinite values NaN # Not a Number
median(c(NA_real_, 2.45, 45.67))
[1] NA
median(c(Inf, 2.45, 45.67))
[1] 45.67
is.numeric(c(NA_real_, 2.45, 45.67))
[1] TRUE
is.numeric(c(Inf, 2.45, 45.67))
[1] TRUE
is.infinite(c(NA_real_, 2.45, 45.67))
[1] FALSE FALSE FALSE
is.infinite(c(Inf, 2.45, 45.67))
[1] TRUE FALSE FALSE
TRUE is 1FALSE is 0TRUE + TRUE
[1] 2
# 1 + 1 + 0 TRUE + TRUE + FALSE
[1] 2
45 * FALSE
[1] 0
c() is the function for concatenate
4 c(43, 5.6, 2.90)
[1] 4 [1] 43.0 5.6 2.9
convert strings to factors, levels is the dictionary
factor(c("AA", "BB", "AA", "CC"))[1] AA BB AA CC Levels: AA BB CC
very important as it can contain anything
list(f = factor(c("AA", "AA")),
v = c(43, 5.6, 2.90),
s = 4)$f [1] AA AA Levels: AA $v [1] 43.0 5.6 2.9 $s [1] 4
won’t dig into those
matrix(1:4, nrow = 2)
[,1] [,2] [1,] 1 3 [2,] 2 4
data.framesame as list but where all objects must have the same length
data.frame(
f = factor(c("AA", "AA", "BB")),
v = c(43, 5.6, 2.90),
s = rep(4, 3))f v s 1 AA 43.0 4 2 AA 5.6 4 3 BB 2.9 4
vdata.frame(
f = factor(c("AA", "AA", "BB")),
v = c(43, 5.6),
s = rep(4, 3))
Error in data.frame(f = factor(c("AA", "AA", "BB")), v = c(43, 5.6), s = rep(4, : arguments imply differing number of rows: 3, 2# evaluate typeof(2)
[1] "double"
class(2)
[1] "numeric"
mode(2)
[1] "numeric"
# check is.integer(2.34)
[1] FALSE
# check with an actual integer is.integer(2L)
[1] TRUE
# convert
is.character("2.34")
[1] TRUE
as.integer(2.34)
[1] 2
as.character(2.34)
[1] "2.34"
as.numeric("2.34")
[1] 2.34
Vectors are the simplest type of object in R.
print(5)
[1] 5
[1] means we made a numeric vector of length 1. Now look at what the : operator does:
1:30
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 [24] 24 25 26 27 28 29 30
How many elements are in the thing we made here? What does the [24] signify?
Think of vectors as collections of simple things (like numbers) that are ordered. We can create vectors from other vectors using the c function:
c(2, TRUE, "a string")
[1] "2" "TRUE" "a string"
We can use the assignment operator <- to associate a name to our vectors in order to reuse them:
my_vec <- c(3, 4, 1:3) my_vec
[1] 3 4 1 2 3
Rstudio has the built-in shortcut Alt+- for <-
Even if = works also for <-, don’t use it, see why
The following will build a character vector. We know this because the elements are all in “quotes”.
char_vec <- c("dog", "cat", "ape")Now use the c function to combine a length-one vector number of the number 4 with the char_vec. What happens?
c(4, char_vec)
[1] "4" "dog" "cat" "ape"
Notice that the 4 is quoted. R turned it into a character vector and then combined it with char_vec.
All elements in a atomic vector must be of the same type. Otherwise, they are silently coerced.
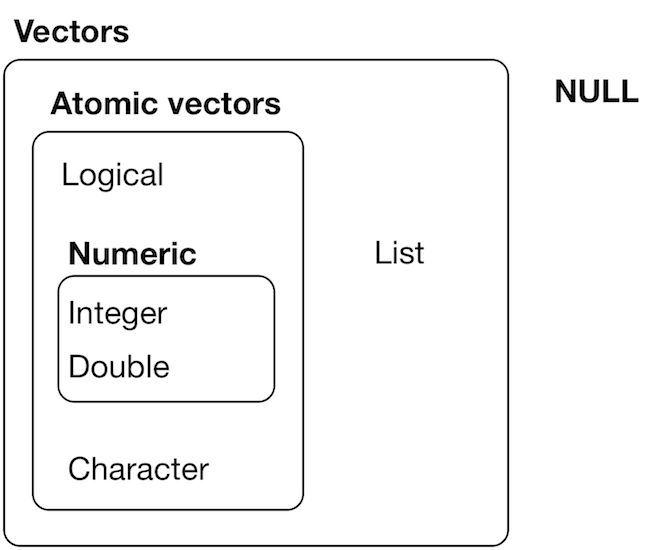
source: H. Wickham - R for data science, licence CC
is.vector(char_vec)
[1] TRUE
is.vector(list(a = 1))
[1] TRUE
is.data.frame(list(a = 1))
[1] FALSE
R has a few built in vectors. One of these is LETTERS. What does it contain?
LETTERS
[1] "A" "B" "C" "D" "E" "F" "G" "H" "I" "J" "K" "L" "M" "N" "O" "P" "Q" [18] "R" "S" "T" "U" "V" "W" "X" "Y" "Z"
How do we extract the first element from this (the letter A)? Here is how to do it:
LETTERS[1]
[1] "A"
Use the square brackets [] to subset vectors
Unlike python or Perl, vectors use 1-based index!!
select elements from position 3 to 10:
LETTERS[3:10]
[1] "C" "D" "E" "F" "G" "H" "I" "J"
: operator does?Take a look:
3:10
[1] 3 4 5 6 7 8 9 10
Can you see how LETTERS[3:10] works now?
[1] "B" "C" "D" "E"
[1] "B" "C" "D" "E" "G"
[1] "A" "B" "C" "D" "E" "Y"
the length of a vector is provided by length()
[1] "B" "C" "D" "E" "F" "G" "H" "I" "J" "K" "L" "M" "N" "O" "P" "Q" "R" [18] "S" "T" "U" "V" "W" "X" "Y" "Z"
subsetting could use negative indexes
indexes from 2 to 5
LETTERS[2:5]
[1] "B" "C" "D" "E"
indexes from 2 to 5 + 7
LETTERS[c(2:5, 7)]
[1] "B" "C" "D" "E" "G"
indexes from 1 to 5 + last one
LETTERS[c(1:5, length(LETTERS) - 1)]
[1] "A" "B" "C" "D" "E" "Y"
indexes except 1
LETTERS[-1]
[1] "B" "C" "D" "E" "F" "G" "H" "I" "J" "K" "L" "M" "N" "O" "P" "Q" "R" [18] "S" "T" "U" "V" "W" "X" "Y" "Z"
Like the dict in python or associative array in Perl, characters can be used as indexes
char_vec[1]
[1] "dog"
names(char_vec) <- c("first", "second", "third")
char_vec["first"]
first "dog"
char_vec[c("first", "third")]
first third "dog" "ape"
char_vec
first second third "dog" "cat" "ape"
the [1] is no longer displayed
LETTERS vector a new name vecletters vector as names for vecvec for the name "m", we don’t need the indicevec <- LETTERS names(vec) <- letters vec["m"]
m "M"
my_vec <- 10:18 my_vec + 2
[1] 12 13 14 15 16 17 18 19 20
R recycles vectors that are too short, without any warnings:
1:10 + c(1, 2)
[1] 2 4 4 6 6 8 8 10 10 12
my_vec * c(1:3)
c(1:3) + c(1:2) * c(1:4)
Warning in c(1:3) + c(1:2) * c(1:4): longer object length is not a multiple of shorter object length
[1] 2 6 6 9
Steps R performs behind the scene are:
multiplication first, duplicate 2nd vector to reach length 4
c(1, 2, 3) + (c(1, 2, 1, 2) * c(1, 2, 3, 4))
Warning in c(1, 2, 3) + (c(1, 2, 1, 2) * c(1, 2, 3, 4)): longer object length is not a multiple of shorter object length
[1] 2 6 6 9
add 1st element to first vector to reach length 4
c(1, 2, 3, 1) + c(1, 4, 3, 8)
[1] 2 6 6 9
x <- numeric(10) x[20] <- 1 head(x, 20)
[1] 0 0 0 0 0 0 0 0 0 0 NA NA NA NA NA NA NA NA NA 1
source: Kevin Ushey
Unlike python that will output index out of range, R expand and fill with missing values silently
Vectors with qualitative data
my_f <- factor(c("cytoplasm", "nucleus", "extracellular", "nucleus", "nucleus"))
my_f
[1] cytoplasm nucleus extracellular nucleus nucleus Levels: cytoplasm extracellular nucleus
Actually, data are represented with numbers
str(my_f)
Factor w/ 3 levels "cytoplasm","extracellular",..: 1 3 2 3 3
ids are called levels. Default is alphabetical sorting
levels(my_f)
[1] "cytoplasm" "extracellular" "nucleus"

For moving around those levels, safest way is to use the forcats package
A matrix is a 2D array
M <- matrix(1:6, ncol = 2, nrow = 3) M
[,1] [,2] [1,] 1 4 [2,] 2 5 [3,] 3 6
M <- matrix(1:6, ncol = 2, nrow = 3, byrow = TRUE) M
[,1] [,2] [1,] 1 2 [2,] 3 4 [3,] 5 6
Similar to a matrix but with dimensions \(\geq\) 3D
A <- array(1:24, dim = c(2, 4, 3)) A
, , 1
[,1] [,2] [,3] [,4]
[1,] 1 3 5 7
[2,] 2 4 6 8
, , 2
[,1] [,2] [,3] [,4]
[1,] 9 11 13 15
[2,] 10 12 14 16
, , 3
[,1] [,2] [,3] [,4]
[1,] 17 19 21 23
[2,] 18 20 22 24Also named recursive vectors. Most permissive type, could contain anything and be nested!
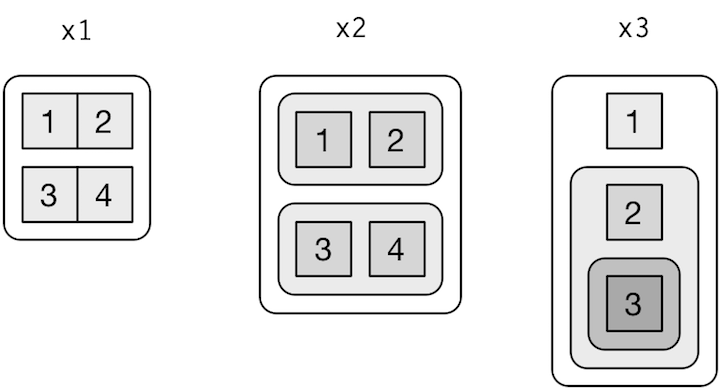
source: H. Wickham - R for data science, licence CC
Indexing lists in #rstats. Inspired by the Residence Inn pic.twitter.com/YQ6axb2w7t
— Hadley Wickham (@hadleywickham) 14 septembre 2015
l <- list(name = "Farina",
firstname = "Geoff",
year = 1995)l["firstname"]
$firstname [1] "Geoff"
l[["firstname"]]
[1] "Geoff"
How to subset a single pepper seed?
It’s the most important type to recall. All the tidyverse is focusing on those.
Actually on tweaked data.frame: tibbles
data.frame are lists where all columns (i.e vectors) are of the same length
women
height weight 1 58 115 2 59 117 3 60 120 4 61 123 5 62 126 6 63 129 7 64 132 8 65 135 9 66 139 10 67 142 11 68 146 12 69 150 13 70 154 14 71 159 15 72 164
We can extract a vector (column) from a data frame in a few different ways:
[[]]women[["height"]]
[1] 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
$ operatorwomen$height
[1] 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72
What would be the output of women["height"]?
A data frame can be considered as a table and extract a specify a cell by its row and column:
head(women, 5)
height weight 1 58 115 2 59 117 3 60 120 4 61 123 5 62 126
[]women[4, 2]
[1] 123
In addition to the arithmetic operators
== equal!= different< smaller<= smaller or equal> greater>= greater or equal! is not&, && and|, || orvec <- c(1, 5, 7) # Example add 1 to each element of vec
res <- vector("numeric", length = length(vec))
for (i in seq_along(vec)) {
res[i] <- vec[i] + 1
}
res
[1] 2 6 8
purrrmap_dbl(vec, ~ .x + 1)
[1] 2 6 8
vec + 1
[1] 2 6 8
write_tsv(mtcars, here::here("results", "mtcars_file.tsv"))file data/mtcars_file.tsv
data/mtcars_file.tsv: cannot open `data/mtcars_file.tsv' (No such file or directory)
write_rds(mtcars, here::here("results", "mtcars_object.rds"))file data/mtcars_object.rds
data/mtcars_object.rds: cannot open `data/mtcars_object.rds' (No such file or directory)
mpg_wt <- lm(mpg ~ wt, data = mtcars) mpg_wt
Call:
lm(formula = mpg ~ wt, data = mtcars)
Coefficients:
(Intercept) wt
37.285 -5.344
typeof(mpg_wt)
[1] "list"
write_rds(mpg_wt, "data/mpg_wt.rds")
mt_lm <- read_rds("data/mpg_wt.rds")
mt_lm
Call:
lm(formula = mpg ~ wt, data = mtcars)
Coefficients:
(Intercept) wt
37.285 -5.344
identical(mpg_wt, mt_lm)
[1] TRUE

Next step is to learn programming!