September 2019
Rmarkdown
Learning objectives
You will learn to:

- use the markdown syntax
- create Rmarkdown documents
- define the output format you expect to render
- use the interactive RStudio interface to
- create your documents
- insert R code
- build your final document
RMarkdown
Why using rmarkdown?
- write detailed reports
- ensure reproducibility
- keep track of your analyses
- comment/describe each step of your analysis
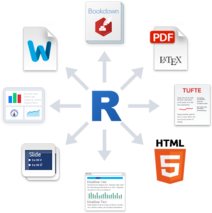
- export a single (Rmd) document to various formats (Pdf, Html…)
- text file that can be managed by a version control system (like git)
Rmarkdown
 +
+  +
+ 
Markdown
Markdown is used to format the text
Markup language
- Such as Xml, HTML
- A coding system used to structure text
- Uses markup tags (e.g.
<h1></h1>in HTML)
HTML
<!DOCTYPE html> <html> <body> <h1>This is a heading</h1> <p>This is some text in a paragraph.</p> </body> </html>
Lightweight markup language
- Easy to read and write as it uses simple tags (e.g.
#)
MD
# This is a heading This is some text in a paragraph
Markdown
common text formatting tags
Headers
- Levels are defined using
#,##,###…
Text style
- bold (
**This will be bold**) - italic (
*This will be italic*)
Links and images
http://example.comis auto-linked[description](http://example.com)
Verbatim code
code(inline coding stuff)- triple backticks are delimiting code blocks
``` This is *verbatim* code # Even headers are not interpreted ```
Rmarkdown cheatsheet

- Have a look at the online documents on the Rmarkdown website
- Use the Cheatsheet in the
Help > Cheatsheetsmenu.
Exercise
Markdown
10:00
Learn to use the markdown syntax

Before writing your own Rmarkdown document, use the excellent ressource on commonmark.org to learn the basics of markdown formatting.
An alternative online ressource can be found on www.markdowntutorial.com
Including R code
Rmarkdown document

from the Rmarkdown cheatsheet
Rmarkdown

- extends markdown
- place R code in chunks
- chunks will be evaluated
- can also handle bash; python; css; …
Knitr

- extracts R chunks
- interprets them
- formats results as markdown
- reintegrates them into the main document (md)
Pandoc
- pandoc converts markdown to the desired document (Pdf, Html, …)
Organising files
Use projects
The only two things that make @JennyBryan 😤😠🤯. Instead use projects + here::here() #rstats pic.twitter.com/GwxnHePL4n
— Hadley Wickham (@hadleywickham) December 11, 2017
Rstudio projects
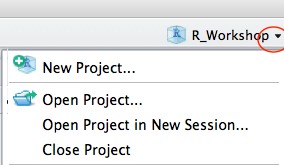
Jennifer Bryan’s advice
Use here package to build paths
- gets the root path of your project:
- detects Rstudio projects (
.Rproj) - git repository (
.git) .herefile
- detects Rstudio projects (
source: Jennifer Bryan’s article and test repo
RStudio projects
create
- a new project
- in a new folder
- no
git
- create a sub-folder
data - test
here::here() - test
here::dr_here()
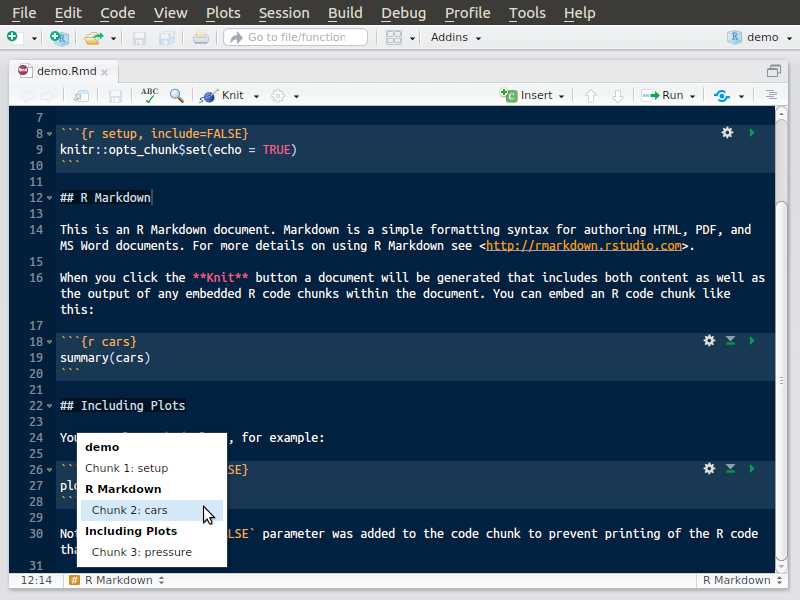
Rmarkdown document
Create, step 1

Rmarkdown document
Create, step 2
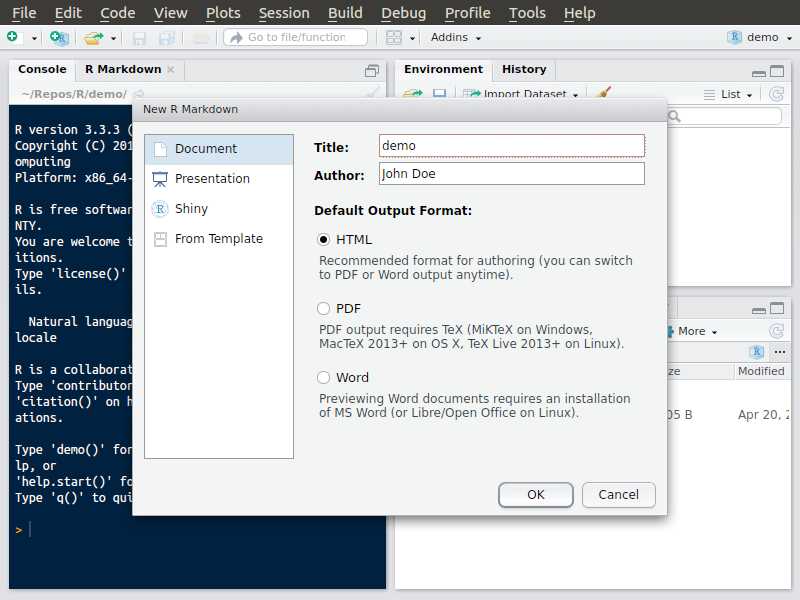
Rmarkdown document
Create, step 3
Generate your first HTML file
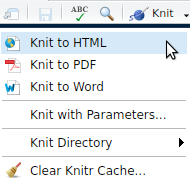
Use the knit button in RStudio
Rmarkdown document
Structure

YAML header
- to define document wide options
- title, name, …
markdown
- markdown syntax to write your descriptions, remarks
- litterate programming
chunks
- code to be interpreted by R
R code chunks
Insert a chunk

R code chunks

- delimited by triple backticks tags (
```) - options in curly brackets
- engine evaluating the code
R but also python, bash, … ```{r}is the minimum to define a starting R chunk- name of chunk
- show or hide the source code
- evaluate it or not
- figure size …
- engine evaluating the code
R code chunks
Inline R code
Integrate small pieces of R code

Use backticks ( ) followed by the keyword r:\ ``r
Example
Type in 1 + 1 = `r 1+1` to render 1 + 1 = 2.
Rmarkdown document
Generate the output document
- use the integrated Knit button.
- call
rmarkdown::render()

Popular output formats
HTML
- fast rendering
- by default embeds binaries (pictures etc.)
- single file
- preserves original aspect
- requires \(\LaTeX\)
have a look at the new TinyTeX package
Word document
- widely used
- easily editable
- collaborate with people not using Rmarkdown
- prepare scientific manuscripts suitable for submission
Bibliography
Supported formats
- Use it with your EndNote or Zotero database:
- BibLaTeX, BibTeX, EndNote, EndNote XML, MEDLINE, ISI, MODS, RIS, Copac, JSON citeproc
Styles
- uses citation style language (
csl) files - have a look at:
Bibliography
How to
- setup in the yaml header
- insert citations using the pandoc syntax:
[@citation-key]
Example
--- title: "Sample Document" output: html_document bibliography: bibliography.bib csl: nature.csl --- Insert your reference [@my-reference] like I did.
Zotero

- install the Better Bib(La)TeX plugin
- adjust the preferences (for better integration)
- export your database as bibtex
- drag and drop pandoc keys to your Rmarkdown document
Import data using readr
Learning objectives
You will learn to:

- use
readrto import your data into R - use the interactive RStudio interface to visualise your data
- appreciate tibbles
Importing data
- Represents probably the first step of your work
- R can handle multiple data types
- flat files (
.csv,.tsv, …) - excel files (
.xls,.xlsx) - foreign statistical formats (
.sasfrom SAS,.savfrom SPSS,.dtafrom Stata) - databases (SQL, SQLite …)
- flat files (
Tidyverse implementation

- R base already provides functions for text files (i.e.
read.csv(),read.delim()) - tidyverse redefines these functions:
- speed
- characters are not coerced to factors by default
- generates tibbles
Tibbles
Tibbles

- have a refined print method that shows only the first 10 rows.
- show all the columns that fit on screen and list the name of remaining ones.
- each column reports its type.
- makes it much easier to work with large data.
Hint

Use as_tibble() to convert a data.frame to a tibble
Tibbles
tibble vs data.frame
data.frame
iris
Sepal.Length Sepal.Width Petal.Length Petal.Width Species 1 5.1 3.5 1.4 0.2 setosa 2 4.9 3.0 1.4 0.2 setosa 3 4.7 3.2 1.3 0.2 setosa 4 4.6 3.1 1.5 0.2 setosa 5 5.0 3.6 1.4 0.2 setosa 6 5.4 3.9 1.7 0.4 setosa 7 4.6 3.4 1.4 0.3 setosa 8 5.0 3.4 1.5 0.2 setosa 9 4.4 2.9 1.4 0.2 setosa 10 4.9 3.1 1.5 0.1 setosa 11 5.4 3.7 1.5 0.2 setosa 12 4.8 3.4 1.6 0.2 setosa 13 4.8 3.0 1.4 0.1 setosa 14 4.3 3.0 1.1 0.1 setosa 15 5.8 4.0 1.2 0.2 setosa 16 5.7 4.4 1.5 0.4 setosa 17 5.4 3.9 1.3 0.4 setosa 18 5.1 3.5 1.4 0.3 setosa 19 5.7 3.8 1.7 0.3 setosa 20 5.1 3.8 1.5 0.3 setosa 21 5.4 3.4 1.7 0.2 setosa 22 5.1 3.7 1.5 0.4 setosa 23 4.6 3.6 1.0 0.2 setosa 24 5.1 3.3 1.7 0.5 setosa 25 4.8 3.4 1.9 0.2 setosa 26 5.0 3.0 1.6 0.2 setosa 27 5.0 3.4 1.6 0.4 setosa 28 5.2 3.5 1.5 0.2 setosa 29 5.2 3.4 1.4 0.2 setosa 30 4.7 3.2 1.6 0.2 setosa 31 4.8 3.1 1.6 0.2 setosa 32 5.4 3.4 1.5 0.4 setosa 33 5.2 4.1 1.5 0.1 setosa 34 5.5 4.2 1.4 0.2 setosa 35 4.9 3.1 1.5 0.2 setosa 36 5.0 3.2 1.2 0.2 setosa 37 5.5 3.5 1.3 0.2 setosa 38 4.9 3.6 1.4 0.1 setosa 39 4.4 3.0 1.3 0.2 setosa 40 5.1 3.4 1.5 0.2 setosa 41 5.0 3.5 1.3 0.3 setosa 42 4.5 2.3 1.3 0.3 setosa 43 4.4 3.2 1.3 0.2 setosa 44 5.0 3.5 1.6 0.6 setosa 45 5.1 3.8 1.9 0.4 setosa 46 4.8 3.0 1.4 0.3 setosa 47 5.1 3.8 1.6 0.2 setosa 48 4.6 3.2 1.4 0.2 setosa 49 5.3 3.7 1.5 0.2 setosa 50 5.0 3.3 1.4 0.2 setosa 51 7.0 3.2 4.7 1.4 versicolor 52 6.4 3.2 4.5 1.5 versicolor 53 6.9 3.1 4.9 1.5 versicolor 54 5.5 2.3 4.0 1.3 versicolor 55 6.5 2.8 4.6 1.5 versicolor 56 5.7 2.8 4.5 1.3 versicolor 57 6.3 3.3 4.7 1.6 versicolor 58 4.9 2.4 3.3 1.0 versicolor 59 6.6 2.9 4.6 1.3 versicolor 60 5.2 2.7 3.9 1.4 versicolor 61 5.0 2.0 3.5 1.0 versicolor 62 5.9 3.0 4.2 1.5 versicolor 63 6.0 2.2 4.0 1.0 versicolor 64 6.1 2.9 4.7 1.4 versicolor 65 5.6 2.9 3.6 1.3 versicolor 66 6.7 3.1 4.4 1.4 versicolor 67 5.6 3.0 4.5 1.5 versicolor 68 5.8 2.7 4.1 1.0 versicolor 69 6.2 2.2 4.5 1.5 versicolor 70 5.6 2.5 3.9 1.1 versicolor 71 5.9 3.2 4.8 1.8 versicolor 72 6.1 2.8 4.0 1.3 versicolor 73 6.3 2.5 4.9 1.5 versicolor 74 6.1 2.8 4.7 1.2 versicolor 75 6.4 2.9 4.3 1.3 versicolor 76 6.6 3.0 4.4 1.4 versicolor 77 6.8 2.8 4.8 1.4 versicolor 78 6.7 3.0 5.0 1.7 versicolor 79 6.0 2.9 4.5 1.5 versicolor 80 5.7 2.6 3.5 1.0 versicolor 81 5.5 2.4 3.8 1.1 versicolor 82 5.5 2.4 3.7 1.0 versicolor 83 5.8 2.7 3.9 1.2 versicolor 84 6.0 2.7 5.1 1.6 versicolor 85 5.4 3.0 4.5 1.5 versicolor 86 6.0 3.4 4.5 1.6 versicolor 87 6.7 3.1 4.7 1.5 versicolor 88 6.3 2.3 4.4 1.3 versicolor 89 5.6 3.0 4.1 1.3 versicolor 90 5.5 2.5 4.0 1.3 versicolor 91 5.5 2.6 4.4 1.2 versicolor 92 6.1 3.0 4.6 1.4 versicolor 93 5.8 2.6 4.0 1.2 versicolor 94 5.0 2.3 3.3 1.0 versicolor 95 5.6 2.7 4.2 1.3 versicolor 96 5.7 3.0 4.2 1.2 versicolor 97 5.7 2.9 4.2 1.3 versicolor 98 6.2 2.9 4.3 1.3 versicolor 99 5.1 2.5 3.0 1.1 versicolor 100 5.7 2.8 4.1 1.3 versicolor 101 6.3 3.3 6.0 2.5 virginica 102 5.8 2.7 5.1 1.9 virginica 103 7.1 3.0 5.9 2.1 virginica 104 6.3 2.9 5.6 1.8 virginica 105 6.5 3.0 5.8 2.2 virginica 106 7.6 3.0 6.6 2.1 virginica 107 4.9 2.5 4.5 1.7 virginica 108 7.3 2.9 6.3 1.8 virginica 109 6.7 2.5 5.8 1.8 virginica 110 7.2 3.6 6.1 2.5 virginica 111 6.5 3.2 5.1 2.0 virginica 112 6.4 2.7 5.3 1.9 virginica 113 6.8 3.0 5.5 2.1 virginica 114 5.7 2.5 5.0 2.0 virginica 115 5.8 2.8 5.1 2.4 virginica 116 6.4 3.2 5.3 2.3 virginica 117 6.5 3.0 5.5 1.8 virginica 118 7.7 3.8 6.7 2.2 virginica 119 7.7 2.6 6.9 2.3 virginica 120 6.0 2.2 5.0 1.5 virginica 121 6.9 3.2 5.7 2.3 virginica 122 5.6 2.8 4.9 2.0 virginica 123 7.7 2.8 6.7 2.0 virginica 124 6.3 2.7 4.9 1.8 virginica 125 6.7 3.3 5.7 2.1 virginica 126 7.2 3.2 6.0 1.8 virginica 127 6.2 2.8 4.8 1.8 virginica 128 6.1 3.0 4.9 1.8 virginica 129 6.4 2.8 5.6 2.1 virginica 130 7.2 3.0 5.8 1.6 virginica 131 7.4 2.8 6.1 1.9 virginica 132 7.9 3.8 6.4 2.0 virginica 133 6.4 2.8 5.6 2.2 virginica 134 6.3 2.8 5.1 1.5 virginica 135 6.1 2.6 5.6 1.4 virginica 136 7.7 3.0 6.1 2.3 virginica 137 6.3 3.4 5.6 2.4 virginica 138 6.4 3.1 5.5 1.8 virginica 139 6.0 3.0 4.8 1.8 virginica 140 6.9 3.1 5.4 2.1 virginica 141 6.7 3.1 5.6 2.4 virginica 142 6.9 3.1 5.1 2.3 virginica 143 5.8 2.7 5.1 1.9 virginica 144 6.8 3.2 5.9 2.3 virginica 145 6.7 3.3 5.7 2.5 virginica 146 6.7 3.0 5.2 2.3 virginica 147 6.3 2.5 5.0 1.9 virginica 148 6.5 3.0 5.2 2.0 virginica 149 6.2 3.4 5.4 2.3 virginica 150 5.9 3.0 5.1 1.8 virginica
Tibbles
tibble vs data.frame
tibble
# library(tibble) as_tibble(iris)
# A tibble: 150 x 5
Sepal.Length Sepal.Width Petal.Length Petal.Width Species
<dbl> <dbl> <dbl> <dbl> <fct>
1 5.1 3.5 1.4 0.2 setosa
2 4.9 3 1.4 0.2 setosa
3 4.7 3.2 1.3 0.2 setosa
4 4.6 3.1 1.5 0.2 setosa
5 5 3.6 1.4 0.2 setosa
6 5.4 3.9 1.7 0.4 setosa
7 4.6 3.4 1.4 0.3 setosa
8 5 3.4 1.5 0.2 setosa
9 4.4 2.9 1.4 0.2 setosa
10 4.9 3.1 1.5 0.1 setosa
# … with 140 more rows
tibble adjusts to width
# A tibble: 150 x 5
Sepal.Length Sepal.Width
<dbl> <dbl>
1 5.1 3.5
2 4.9 3
3 4.7 3.2
4 4.6 3.1
5 5 3.6
6 5.4 3.9
7 4.6 3.4
8 5 3.4
9 4.4 2.9
10 4.9 3.1
# … with 140 more rows, and 3
# more variables:
# Petal.Length <dbl>,
# Petal.Width <dbl>,
# Species <fct>tibble printing enhancements
- column type is visible
- shows only the first 10 rows
- shows only the columns that fit on the screen
Create tibbles
tibble()
- similar to
base::data.frame()but- does not coerce characters to factors
- does not change column names
- never uses rownames
data.frame(`bad name` = 1:4,
x = rep(letters[1:2], 2)) %>%
str()'data.frame': 4 obs. of 2 variables: $ bad.name: int 1 2 3 4 $ x : Factor w/ 2 levels "a","b": 1 2 1 2
tibble(`bad name` = 1:4,
x = rep(letters[1:2], 2)) %>%
str()Classes 'tbl_df', 'tbl' and 'data.frame': 4 obs. of 2 variables: $ bad name: int 1 2 3 4 $ x : chr "a" "b" "a" "b"
The tidyverse packages to import your data
Tidyverse packages to import your data

Seven file formats are supported by the readr package:
read_csv(): comma separated (CSV) filesread_tsv(): tab separated filesread_delim(): general delimited filesread_fwf(): fixed width filesread_table(): tabular files where colums are separated by white-space.read_log(): web log files
readxl

To import excel files (.xls and .xlsx):
read_excel()read_xls()read_xlsx()
haven

read_sas()for SASread_sav()for SPSSread_dta()for Stata
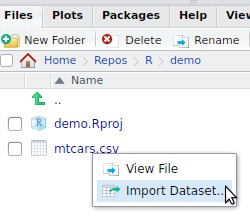
Importing flat files
Reading flat files
Flat file example: mtcars.csv
- Create a new project (finding your files will be easier)
- Download the
mtcars.csvfile to your project folder (using your favourite browser) - Open the file with a text viewer and have a look at its content
"mpg","cyl","disp","hp","drat","wt","qsec","vs","am","gear","carb" 21,6,160,110,3.9,2.62,16.46,0,1,4,4 21,6,160,110,3.9,2.875,17.02,0,1,4,4 22.8,4,108,93,3.85,2.32,18.61,1,1,4,1 21.4,6,258,110,3.08,3.215,19.44,1,0,3,1 18.7,8,360,175,3.15,3.44,17.02,0,0,3,2 ...
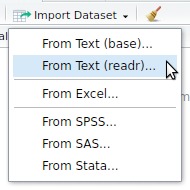
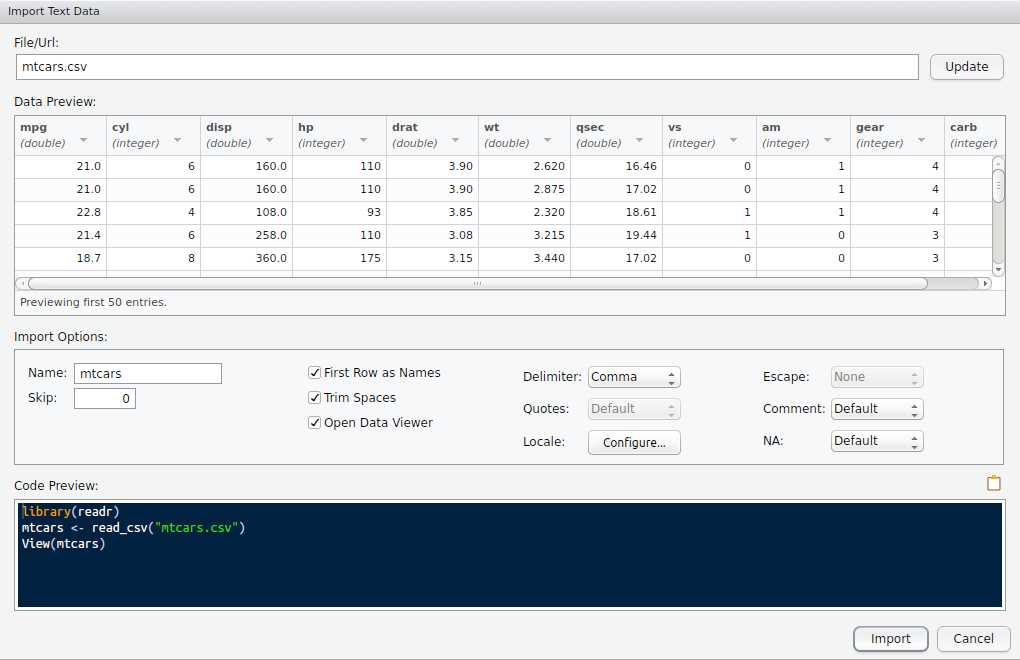
Rstudio data import
interactive call to readr
Exercise
Import the mtcars.csv dataset
- Use the interactive
Import Datasetbutton to import themtcars.csvfile.
Rstudio data import
preview window
Reading flat files
comma separated values
read_csv()
- to import comma separated values
- is able to read local and remote files
- is able to read compressed files (
.zip,.gz, …)
Reading flat files
comma separated values
Using read_csv()
read_csv(here::here("data", "mtcars.csv"))Parsed with column specification: cols( mpg = col_double(), cyl = col_double(), disp = col_double(), hp = col_double(), drat = col_double(), wt = col_double(), qsec = col_double(), vs = col_double(), am = col_double(), gear = col_double(), carb = col_double() )
# A tibble: 32 x 11
mpg cyl disp hp drat wt qsec vs am gear carb
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 21 6 160 110 3.9 2.62 16.5 0 1 4 4
2 21 6 160 110 3.9 2.88 17.0 0 1 4 4
3 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
4 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1
5 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2
6 18.1 6 225 105 2.76 3.46 20.2 1 0 3 1
7 14.3 8 360 245 3.21 3.57 15.8 0 0 3 4
8 24.4 4 147. 62 3.69 3.19 20 1 0 4 2
9 22.8 4 141. 95 3.92 3.15 22.9 1 0 4 2
10 19.2 6 168. 123 3.92 3.44 18.3 1 0 4 4
# … with 22 more rowsColumn types
Column types
- are guessed from the 1000 first rows
- adjustable
guess_maxoption
- adjustable
- guessed types are displayed as a message
- to hide this message:
- lazy method 1: set
message = FALSEin your rmarkdown chunk option. - lazy method 2: set
col_types = cols() - Hadley Wickham recommends to adjust the
col_typesto avoid any problem
- lazy method 1: set
Message
Parsed with column specification: cols( mpg = col_double(), cyl = col_double(), disp = col_double(), hp = col_double(), drat = col_double(), wt = col_double(), qsec = col_double(), vs = col_double(), am = col_double(), gear = col_double(), carb = col_double() )
Column types
The col_types argument
exa <- here::here("data", "example.csv")read_csv(exa, col_types = cols())
# A tibble: 3 x 3 animal colour value <chr> <chr> <dbl> 1 dog red 1 2 cat blue 2 3 chicken green 6
- Let’s start with a file containing only 3 columns:
animal,colourandvalue - Column types are specified using the
cols()function - Types can be one of
double,integer,character,logical,factor,date,datetimeortime
Column types
Explicit method
Using a function defining each type:
col_double()col_integer()col_character()col_logical()col_factor()col_date()col_datetime()col_time()
Or telling to guess or skip a column:
col_guess()col_skip()
Example
read_csv(exa,
col_types = cols(
animal = col_character(),
colour = col_character(),
value = col_integer()
))
# A tibble: 3 x 3 animal colour value <chr> <chr> <int> 1 dog red 1 2 cat blue 2 3 chicken green 6
Column types
Compact shortcuts
Using a single character to define each type:
c= characteri= integern= numberd= doublel= logicalD= dateT= date timet= time
Or telling to guess or skip a column:
?= guess_or-= skip
Example
read_csv(exa,
col_types = cols(
animal = "c",
colour = "c",
value = "i"
))
# A tibble: 3 x 3 animal colour value <chr> <chr> <int> 1 dog red 1 2 cat blue 2 3 chicken green 6
Even more compact
read_csv(exa, col_types = "cci")
- Use the single character code in the order of appearance of each column
Exercise
Override the detected column types
- import the
example.csvfile but- skip the
colourcolumn - read in the
valuecolumn as double
- skip the
Exercise
Answer
read_csv(exa, col_types = cols(animal = col_character(),
colour = col_skip(),
value = col_double()))# A tibble: 3 x 2 animal value <chr> <dbl> 1 dog 1 2 cat 2 3 chicken 6
read_csv(exa, col_types = cols(animal = "c",
colour = "_",
value = "d"))# A tibble: 3 x 2 animal value <chr> <dbl> 1 dog 1 2 cat 2 3 chicken 6
read_csv(exa, col_types = "c_d")
# A tibble: 3 x 2 animal value <chr> <dbl> 1 dog 1 2 cat 2 3 chicken 6
Column names
The col_names argument
- can be either
TRUE,FALSEor a character vector. - default value is
TRUE - if
TRUE, the first row will be used as column names - if
FALSE, names are generated (X1, X2, X3, …) - if it is a character vector, it will define the column names
Example
read_csv(exa,
col_names = TRUE)
# A tibble: 3 x 3 animal colour value <chr> <chr> <dbl> 1 dog red 1 2 cat blue 2 3 chicken green 6
read_csv(exa,
col_names = FALSE)
# A tibble: 4 x 3 X1 X2 X3 <chr> <chr> <chr> 1 animal colour value 2 dog red 1 3 cat blue 2 4 chicken green 6
read_csv(exa,
col_names = c("name", "colname", "number"))
# A tibble: 4 x 3 name colname number <chr> <chr> <chr> 1 animal colour value 2 dog red 1 3 cat blue 2 4 chicken green 6
Column names
Hint

col_namesis handy if they are no column names in the file- If you would like to rename columns, use
dplyr::rename()(see upcomingdplyrlecture).
read_csv(exa, col_names = c("name", "colname", "number"))
# A tibble: 4 x 3 name colname number <chr> <chr> <chr> 1 animal colour value 2 dog red 1 3 cat blue 2 4 chicken green 6
read_csv(exa, col_names = TRUE) %>%
rename(name = animal,
colname = colour,
number = value)
# A tibble: 3 x 3 name colname number <chr> <chr> <dbl> 1 dog red 1 2 cat blue 2 3 chicken green 6
Skipping lines
skip argument
To skip the first n rows
n_max argument
To stop reading after n rows
Hint

You might want to adjust col_names to get what you want
readr_example("mtcars.csv") %>%
read_csv(skip = 3,
n_max = 3,
col_names = FALSE)# A tibble: 3 x 11
X1 X2 X3 X4 X5 X6 X7 X8 X9 X10 X11
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
2 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1
3 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2readr_example("mtcars.csv") %>%
read_csv(skip = 3, n_max = 3,
col_names = c("mpg", "cyl", "disp", "hp", "drat", "wt", "qsec", "vs", "am", "gear", "carb"))# A tibble: 3 x 11
mpg cyl disp hp drat wt qsec vs am gear carb
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
2 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1
3 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2readr functions
read_csv()
- Comma delimited files
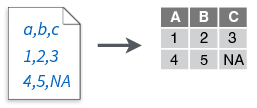
read_csv2()
- Semi-colon delimited files
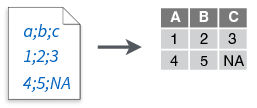
read_tsv()
- tab delimited files

read_delim()
- any delimiter:

read_delim(file, delim = "|", ...)
read_fwf()
- fixed width files

If speed is still an issue
fread from data.table
- stable
install.packages("data.table")- overhauled
- 2X faster than
readr
promising vroom
- on CRAN
install.packages("vroom")- by Jim Hester (
readr) - same syntax!
- multi-threaded
ALTREPframework- 18X faster than
readrin some conditions
Wrap up
You learned to:

- appreciate the tibble printing features
- column types are displayed
- use
readrto import your flat file data into R- using the command line
- using the interactive RStudio interface
- adjust the imported data types
Before we stop
Further reading

- R for Data Science
- http://readr.tidyverse.org/
vignette("readr")
- Datacamp course
Acknowledgments
- Hadley Wickham