Pipeline
September 2019

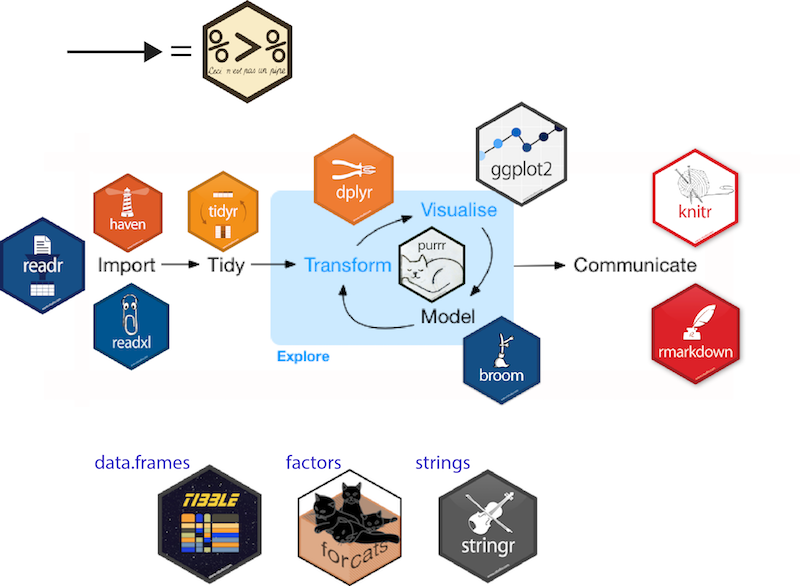
ggplot2, for data visualizationdplyr, for data manipulationtidyr, for data tidyingreadr, for data importpurrr, for functional programmingtibble, for tibbles, a modern re-imagining of data framessource: http://tidyverse.tidyverse.org/. H.Wickham
hms, for timesstringr, for stringslubridate, for date/timesforcats, for factorsfeather, for sharing datahaven, for SPSS, SAS and Stata fileshttr, for web apisjsonlite for JSONreadxl, for .xls and .xlsx filesrvest, for web scrapingxml2, for XML filesmodelr, for modelling within a pipelinebroom, for models -> tidy data@ucfagls yeah. I think the tidyverse is a dialect. But its accent isn’t so thick
— Hadley Wickham (@hadleywickham) 12 janvier 2017

data.table is faster, for less than 10 m rows, negligible.
Realized today: #tidyverse R and base #rstats have little in common. Beware when looking for job which requires knowledge of R.
— Yeedle N. (@Yeedle) 2 mars 2017
tibbles are nice but a lot of non-tidyverse packages require matrices. rownames still an issue.Anyway, learning the tidyverse does not prevent to learn R base, it helps to get things done early in the process
source: rdocumentation (2017/04/18)
source: rdocumentation (2017/04/18)

set.seed(12) round(mean(rnorm(5)), 2)
[1] -0.76
set.seed(12) rnorm(5) %>% mean() %>% round(2)
[1] -0.76
Of note, magrittr needs to loaded with either:
library(magrittr) library(dplyr) library(tidyverse)


Jenny Bryan in Jeff Leek blog post

All happy families are alike; each unhappy family is unhappy in its own way. Leo Tolstoy, Anna Karenina
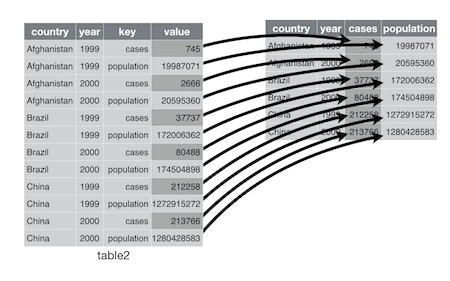
source: Garret Grolemund and vignette("tidy-data")

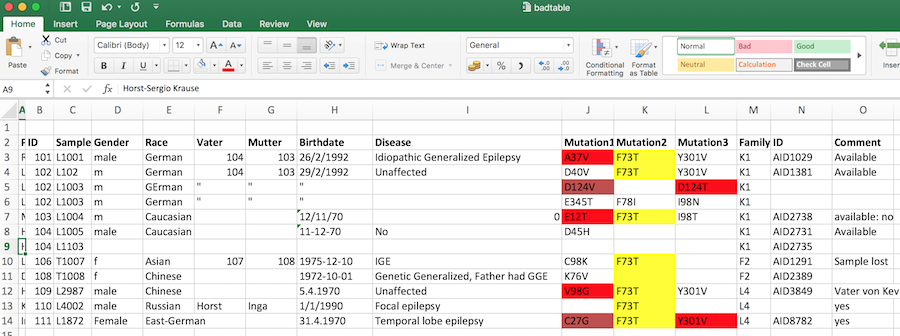
| Error | Tidy violation | Comment |
|---|---|---|
| Patient names | No | Data protection violation |
| Identical column names | Yes | Variable error |
| Inconsistent variables names | No | Bad practice |
| Non-English columns names | No | Bad practice |
| Color coding | No | The horror, the horror |
| Inconsistent dates | No | Use ISO8601 |
| Multiple columns for one item | Yes | One observation per line |
| Redundant information | Yes | Each variable is in its own column |
| Repeated rows | Yes | Each observation is in its own row |
| Uncoded syndromes | Yes/No | Each value in its own cell |
| Unnecessary information | No | like birthdate, comments: bad practice |
| Name of the table | No | You’ll see this often |
10 min, discuss
gather)separate)gather-spread)nest or table)dplyr data transformation)dplyr, combine into single table)
The wide format is generally untidy but found in the majority of datasets

count(mtcars, am, cyl)
# A tibble: 6 x 3
am cyl n
<dbl> <dbl> <int>
1 0 4 3
2 0 6 4
3 0 8 12
4 1 4 8
5 1 6 3
6 1 8 2count(mtcars, am, cyl) %>%
pivot_wider(names_from = cyl,
values_from = n) -> wcars
wcars
# A tibble: 2 x 4
am `4` `6` `8`
<dbl> <int> <int> <int>
1 0 3 4 12
2 1 8 3 2wcars %>%
pivot_longer(cols = -am,
names_to = "cyl",
values_to = "n",
# optional prototype to get integers (not chr)
names_ptypes = list(cyl = integer()))# A tibble: 6 x 3
am cyl n
<dbl> <int> <int>
1 0 4 3
2 0 6 4
3 0 8 12
4 1 4 8
5 1 6 3
6 1 8 2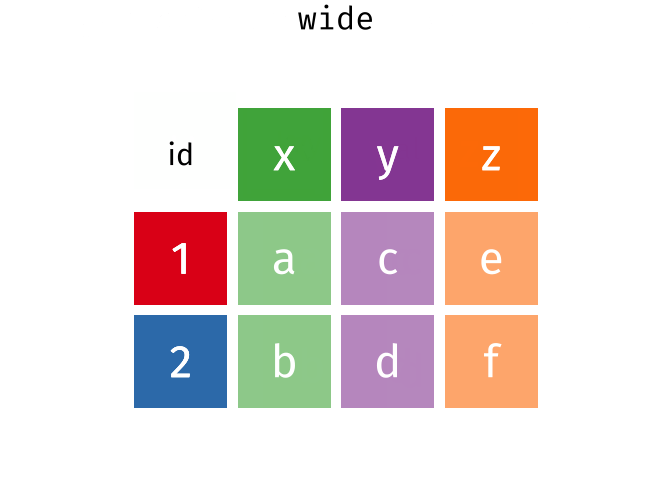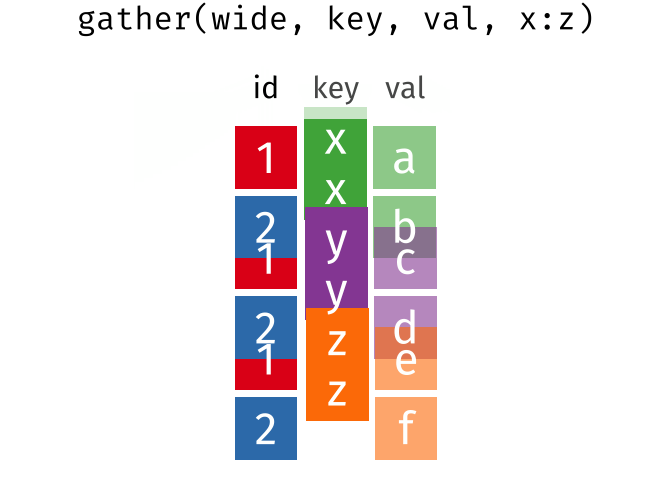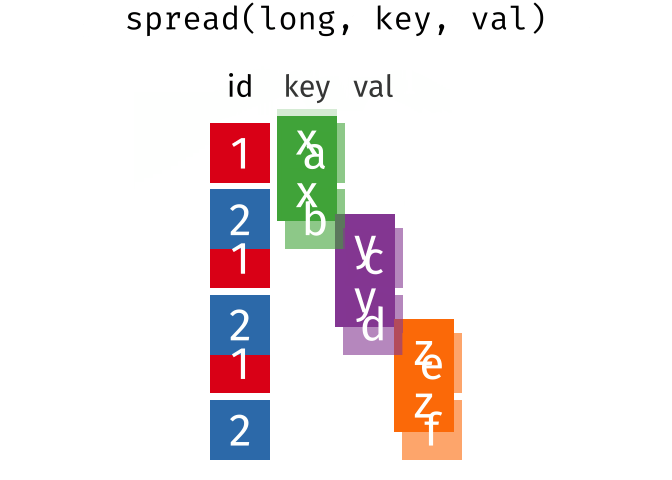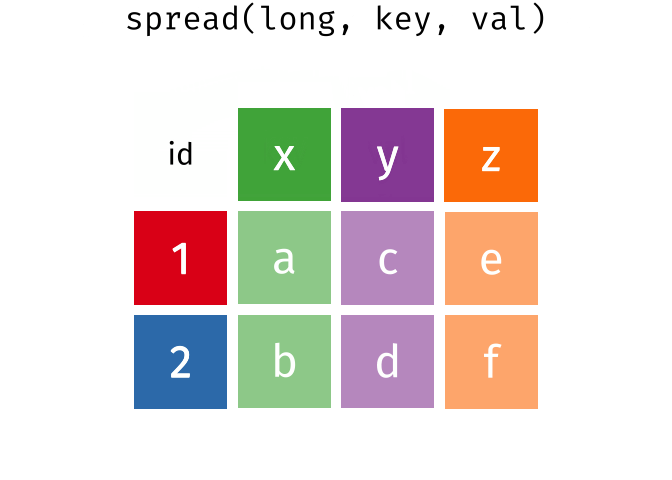
image from Garrick Aden-Buie, github repo
head(fish_encounters, 3)
# A tibble: 3 x 3 fish station seen <fct> <fct> <int> 1 4842 Release 1 2 4842 I80_1 1 3 4842 Lisbon 1
dim(fish_encounters)
[1] 114 3
library(tidyverse)
fish_wide <- fish_encounters %>%
pivot_wider(
names_from = station,
values_from = seen
)
fish_wide
# A tibble: 19 x 12 fish Release I80_1 Lisbon Rstr Base_TD BCE BCW BCE2 BCW2 MAE <fct> <int> <int> <int> <int> <int> <int> <int> <int> <int> <int> 1 4842 1 1 1 1 1 1 1 1 1 1 2 4843 1 1 1 1 1 1 1 1 1 1 3 4844 1 1 1 1 1 1 1 1 1 1 4 4845 1 1 1 1 1 NA NA NA NA NA 5 4847 1 1 1 NA NA NA NA NA NA NA 6 4848 1 1 1 1 NA NA NA NA NA NA 7 4849 1 1 NA NA NA NA NA NA NA NA 8 4850 1 1 NA 1 1 1 1 NA NA NA 9 4851 1 1 NA NA NA NA NA NA NA NA 10 4854 1 1 NA NA NA NA NA NA NA NA 11 4855 1 1 1 1 1 NA NA NA NA NA 12 4857 1 1 1 1 1 1 1 1 1 NA 13 4858 1 1 1 1 1 1 1 1 1 1 14 4859 1 1 1 1 1 NA NA NA NA NA 15 4861 1 1 1 1 1 1 1 1 1 1 16 4862 1 1 1 1 1 1 1 1 1 NA 17 4863 1 1 NA NA NA NA NA NA NA NA 18 4864 1 1 NA NA NA NA NA NA NA NA 19 4865 1 1 1 NA NA NA NA NA NA NA # … with 1 more variable: MAW <int>
fish_wide %>%
pivot_longer(
cols = -fish,
names_to = "station",
values_to = "seen"
)
# A tibble: 209 x 3 fish station seen <fct> <chr> <int> 1 4842 Release 1 2 4842 I80_1 1 3 4842 Lisbon 1 4 4842 Rstr 1 5 4842 Base_TD 1 6 4842 BCE 1 7 4842 BCW 1 8 4842 BCE2 1 9 4842 BCW2 1 10 4842 MAE 1 # … with 199 more rows

Note that we get more rows than in the original dataset, as missing combination are now NA
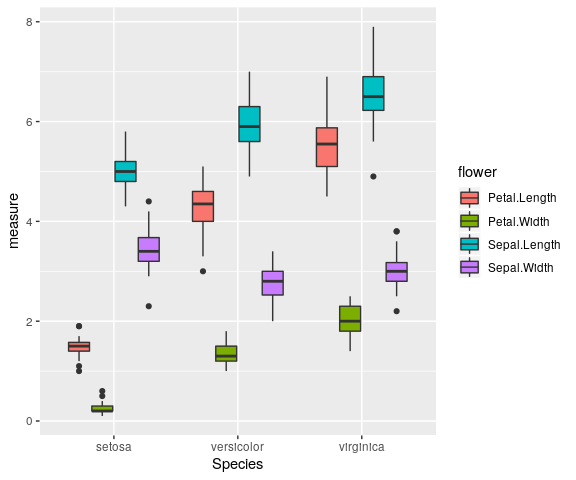
iris %>%
pivot_longer(cols = -Species,
names_to = "flower",
values_to = "measure") %>%
ggplot() +
geom_boxplot(aes(x = Species, y = measure, fill = flower))
demo_tibble <- tibble(year = c(2015, 2014, 2014),
month = c(11L, 2L, 4L), # create a vector of integers
day = c(23, 1, 30), # default is double
value = c("high", "low", "low"))
demo_tibble
# A tibble: 3 x 4 year month day value <dbl> <int> <dbl> <chr> 1 2015 11 23 high 2 2014 2 1 low 3 2014 4 30 low
demo_tibble %>%
unite(date, c(year, month, day),
sep = "-") -> demo_tibble_unite
demo_tibble_unite
# A tibble: 3 x 2 date value <chr> <chr> 1 2015-11-23 high 2 2014-2-1 low 3 2014-4-30 low
use quotes since we are not refering to objects
demo_tibble_unite %>%
separate(date, c("year", "month", "day"))
# A tibble: 3 x 4 year month day value <chr> <chr> <chr> <chr> 1 2015 11 23 high 2 2014 2 1 low 3 2014 4 30 low
patient_df <- tibble(
subject_id = 1001:1003,
visit_id = c("1,2,3", "1,2", "1"),
measured = c("9,0, 11", "11, 3" , "12") )
patient_df
# A tibble: 3 x 3
subject_id visit_id measured
<int> <chr> <chr>
1 1001 1,2,3 9,0, 11
2 1002 1,2 11, 3
3 1003 1 12 Note the incoherent white space
separate_rows(patient_df,
visit_id, measured,
convert = TRUE) # chr -> int
# A tibble: 6 x 3
subject_id visit_id measured
<int> <int> <int>
1 1001 1 9
2 1001 2 0
3 1001 3 11
4 1002 1 11
5 1002 2 3
6 1003 1 12To split single variables use separate
separate() and unite() dummy <- data_frame(year = c(2015, 2014, 2014),
month = c(11, 2, 4),
day = c(23, 1, 30),
value = c("high", "low", "low"))Warning: `data_frame()` is deprecated, use `tibble()`. This warning is displayed once per session.
dummy
# A tibble: 3 x 4 year month day value <dbl> <dbl> <dbl> <chr> 1 2015 11 23 high 2 2014 2 1 low 3 2014 4 30 low
unite()dummy_unite <- unite(dummy, date,
year, month, day,
sep = "-")
dummy_unite# A tibble: 3 x 2 date value <chr> <chr> 1 2015-11-23 high 2 2014-2-1 low 3 2014-4-30 low
separate() and unite() separate()separate(dummy_unite,
date, c("year", "month", "day"))# A tibble: 3 x 4 year month day value <chr> <chr> <chr> <chr> 1 2015 11 23 high 2 2014 2 1 low 3 2014 4 30 low
kelpdf <- data.frame(
Year = c(1999, 2000, 2004, 1999, 2004),
Taxon = c("Saccharina", "Saccharina", "Saccharina", "Agarum", "Agarum"),
Abundance = c(4, 5, 2, 1, 8)
)
kelpdfYear Taxon Abundance 1 1999 Saccharina 4 2 2000 Saccharina 5 3 2004 Saccharina 2 4 1999 Agarum 1 5 2004 Agarum 8
how to fill it?
complete()complete(kelpdf,
Year, Taxon)
# A tibble: 6 x 3 Year Taxon Abundance <dbl> <fct> <dbl> 1 1999 Agarum 1 2 1999 Saccharina 4 3 2000 Agarum NA 4 2000 Saccharina 5 5 2004 Agarum 8 6 2004 Saccharina 2
example from imachorda.com
how to fill out this info with 0s?
complete(), option fillcomplete(kelpdf,
Year, Taxon,
fill = list(Abundance = 0))# A tibble: 6 x 3 Year Taxon Abundance <dbl> <fct> <dbl> 1 1999 Agarum 1 2 1999 Saccharina 4 3 2000 Agarum 0 4 2000 Saccharina 5 5 2004 Agarum 8 6 2004 Saccharina 2
example from imachorda.com
how to fill out this info with 0s?
complete(), option fill and helper full_seq()complete(kelpdf,
# helper tidyr::full_seq
Year = full_seq(Year, period = 1),
Taxon,
fill = list(Abundance = 0))# A tibble: 12 x 3
Year Taxon Abundance
<dbl> <fct> <dbl>
1 1999 Agarum 1
2 1999 Saccharina 4
3 2000 Agarum 0
4 2000 Saccharina 5
5 2001 Agarum 0
6 2001 Saccharina 0
7 2002 Agarum 0
8 2002 Saccharina 0
9 2003 Agarum 0
10 2003 Saccharina 0
11 2004 Agarum 8
12 2004 Saccharina 2example from imachorda.com
separate_rows(patient_df,
visit_id, measured,
convert = TRUE)# A tibble: 6 x 3
subject_id visit_id measured
<int> <int> <int>
1 1001 1 9
2 1001 2 0
3 1001 3 11
4 1002 1 11
5 1002 2 3
6 1003 1 12complete(), with helper nesting()patient_complete <- separate_rows(patient_df,
visit_id, measured,
convert = TRUE) %>%
complete(subject_id, nesting(visit_id))
patient_complete# A tibble: 9 x 3
subject_id visit_id measured
<int> <int> <int>
1 1001 1 9
2 1001 2 0
3 1001 3 11
4 1002 1 11
5 1002 2 3
6 1002 3 NA
7 1003 1 12
8 1003 2 NA
9 1003 3 NAnest()patient_nested <- patient_complete %>% nest(visit_id, measured)
Warning: All elements of `...` must be named. Did you want `data = c(visit_id, measured)`?
patient_nested
# A tibble: 3 x 2
subject_id data
<int> <list<df[,2]>>
1 1001 [3 × 2]
2 1002 [3 × 2]
3 1003 [3 × 2]unnest()unnest(patient_nested)
Warning: `cols` is now required. Please use `cols = c(data)`
# A tibble: 9 x 3
subject_id visit_id measured
<int> <int> <int>
1 1001 1 9
2 1001 2 0
3 1001 3 11
4 1002 1 11
5 1002 2 3
6 1002 3 NA
7 1003 1 12
8 1003 2 NA
9 1003 3 NAgroup_by and nest()patient_complete %>% group_by(subject_id) %>% nest(.key = "visit")
Warning: `.key` is deprecated
# A tibble: 3 x 2
# Groups: subject_id [3]
subject_id visit
<int> <list<df[,2]>>
1 1001 [3 × 2]
2 1002 [3 × 2]
3 1003 [3 × 2]data by defaulttidyr
complete, separate_rows and nest()base to tidyverse, Rajesh Korde, blog’ post
Comments
dplyrtidyranddplyrare intertwinedtidyrways